
In digital image processing and computer vision, image segmentation is the process of partitioning a digital image into multiple image segments, also known as image regions or image objects. The goal of segmentation is to simplify and/or change the representation of an image into something that is more meaningful and easier to analyze. Image segmentation is typically used to locate objects and boundaries in images. More precisely, image segmentation is the process of assigning a label to every pixel in an image such that pixels with the same label share certain characteristics.
An autoencoder is a type of artificial neural network used to learn efficient codings of unlabeled data. An autoencoder learns two functions: an encoding function that transforms the input data, and a decoding function that recreates the input data from the encoded representation. The autoencoder learns an efficient representation (encoding) for a set of data, typically for dimensionality reduction.

Deep learning is the subset of machine learning methods based on artificial neural networks (ANNs) with representation learning. The adjective "deep" refers to the use of multiple layers in the network. Methods used can be either supervised, semi-supervised or unsupervised.

The MNIST database is a large database of handwritten digits that is commonly used for training various image processing systems. The database is also widely used for training and testing in the field of machine learning. It was created by "re-mixing" the samples from NIST's original datasets. The creators felt that since NIST's training dataset was taken from American Census Bureau employees, while the testing dataset was taken from American high school students, it was not well-suited for machine learning experiments. Furthermore, the black and white images from NIST were normalized to fit into a 28x28 pixel bounding box and anti-aliased, which introduced grayscale levels.
Convolutional neural network (CNN) is a regularized type of feed-forward neural network that learns feature engineering by itself via filters optimization. Vanishing gradients and exploding gradients, seen during backpropagation in earlier neural networks, are prevented by using regularized weights over fewer connections. For example, for each neuron in the fully-connected layer 10,000 weights would be required for processing an image sized 100 × 100 pixels. However, applying cascaded convolution kernels, only 25 neurons are required to process 5x5-sized tiles. Higher-layer features are extracted from wider context windows, compared to lower-layer features.
Multimodal learning, in the context of machine learning, is a type of deep learning using a combination of various modalities of data, often arising in real-world applications. An example of multi-modal data is data that combines text with imaging data consisting of pixel intensities and annotation tags. As these modalities have fundamentally different statistical properties, combining them is non-trivial, which is why specialized modelling strategies and algorithms are required. The model is then trained to able to understand and work with multiple forms of data.
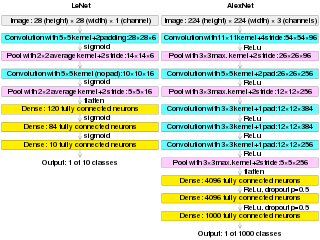
AlexNet is the name of a convolutional neural network (CNN) architecture, designed by Alex Krizhevsky in collaboration with Ilya Sutskever and Geoffrey Hinton, who was Krizhevsky's Ph.D. advisor at the University of Toronto.
Deep image prior is a type of convolutional neural network used to enhance a given image with no prior training data other than the image itself. A neural network is randomly initialized and used as prior to solve inverse problems such as noise reduction, super-resolution, and inpainting. Image statistics are captured by the structure of a convolutional image generator rather than by any previously learned capabilities.
Neural architecture search (NAS) is a technique for automating the design of artificial neural networks (ANN), a widely used model in the field of machine learning. NAS has been used to design networks that are on par or outperform hand-designed architectures. Methods for NAS can be categorized according to the search space, search strategy and performance estimation strategy used:
In computer vision, SqueezeNet is the name of a deep neural network for image classification that was released in 2016. SqueezeNet was developed by researchers at DeepScale, University of California, Berkeley, and Stanford University. In designing SqueezeNet, the authors' goal was to create a smaller neural network with fewer parameters while achieving competitive accuracy.

Neural style transfer (NST) refers to a class of software algorithms that manipulate digital images, or videos, in order to adopt the appearance or visual style of another image. NST algorithms are characterized by their use of deep neural networks for the sake of image transformation. Common uses for NST are the creation of artificial artwork from photographs, for example by transferring the appearance of famous paintings to user-supplied photographs. Several notable mobile apps use NST techniques for this purpose, including DeepArt and Prisma. This method has been used by artists and designers around the globe to develop new artwork based on existent style(s).
Artificial neural networks (ANNs) are models created using machine learning to perform a number of tasks. Their creation was inspired by neural circuitry. While some of the computational implementations ANNs relate to earlier discoveries in mathematics, the first implementation of ANNs was by psychologist Frank Rosenblatt, who developed the perceptron. Little research was conducted on ANNs in the 1970s and 1980s, with the AAAI calling that period an "AI winter".
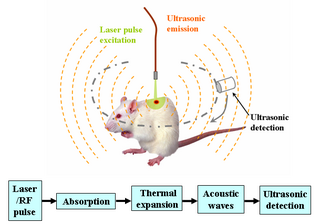
Deep learning in photoacoustic imaging combines the hybrid imaging modality of photoacoustic imaging (PA) with the rapidly evolving field of deep learning. Photoacoustic imaging is based on the photoacoustic effect, in which optical absorption causes a rise in temperature, which causes a subsequent rise in pressure via thermo-elastic expansion. This pressure rise propagates through the tissue and is sensed via ultrasonic transducers. Due to the proportionality between the optical absorption, the rise in temperature, and the rise in pressure, the ultrasound pressure wave signal can be used to quantify the original optical energy deposition within the tissue.
The Fréchet inception distance (FID) is a metric used to assess the quality of images created by a generative model, like a generative adversarial network (GAN). Unlike the earlier inception score (IS), which evaluates only the distribution of generated images, the FID compares the distribution of generated images with the distribution of a set of real images. The FID metric does not completely replace the IS metric. Classifiers that achieve the best (lowest) FID score tend to have greater sample variety while classifiers achieving the best (highest) IS score tend to have better quality within individual images.

Video super-resolution (VSR) is the process of generating high-resolution video frames from the given low-resolution video frames. Unlike single-image super-resolution (SISR), the main goal is not only to restore more fine details while saving coarse ones, but also to preserve motion consistency.
A graph neural network (GNN) belongs to a class of artificial neural networks for processing data that can be represented as graphs.
A vision transformer (ViT) is a transformer designed for computer vision. A ViT breaks down an input image into a series of patches, serialises each patch into a vector, and maps it to a smaller dimension with a single matrix multiplication. These vector embeddings are then processed by a transformer encoder as if they were token embeddings.
Perceiver is a transformer adapted to be able to process non-textual data, such as images, sounds and video, and spatial data. Transformers underlie other notable systems such as BERT and GPT-3, which preceded Perceiver. It adopts an asymmetric attention mechanism to distill inputs into a latent bottleneck, allowing it to learn from large amounts of heterogeneous data. Perceiver matches or outperforms specialized models on classification tasks.
Biomedical data science is a multidisciplinary field which leverages large volumes of data to promote biomedical innovation and discovery. Biomedical data science draws from various fields including Biostatistics, Biomedical informatics, and machine learning, with the goal of understanding biological and medical data. It can be viewed as the study and application of data science to solve biomedical problems. Modern biomedical datasets often have specific features which make their analyses difficult, including:

Stable Diffusion is a deep learning, text-to-image model released in 2022 based on diffusion techniques. It is considered to be a part of the ongoing AI boom.








