This article includes a list of references, related reading or external links, but its sources remain unclear because it lacks inline citations .(May 2014) (Learn how and when to remove this template message) |
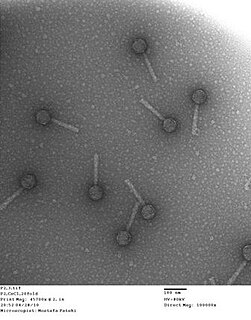
Experimental evolution studies are a means of testing evolutionary theory under carefully designed, reproducible experiments. Given enough time, space, and money, any organism could be used for experimental evolution studies. However, those with rapid generation times, high mutation rates, large population sizes, and small sizes increase the feasibility of experimental studies in a laboratory context. For these reasons, bacteriophages (i.e. viruses that infect bacteria) are especially favored by experimental evolutionary biologists. Bacteriophages, and microbial organisms, can be frozen in stasis, facilitating comparison of evolved strains to ancestors. Additionally, microbes are especially labile from a molecular biologic perspective. Many molecular tools have been developed to manipulate the genetic material of microbial organisms, and because of their small genome sizes, sequencing the full genomes of evolved strains is trivial. Therefore, comparisons can be made for the exact molecular changes in evolved strains during adaptation to novel conditions.
Experimental evolution is the use of laboratory experiments or controlled field manipulations to explore evolutionary dynamics. Evolution may be observed in the laboratory as individuals/populations adapt to new environmental conditions by natural selection. There are two different ways in which adaptation can arise in experimental evolution. One is via an individual organism gaining a novel beneficial mutation. The other is from allele frequency change in standing genetic variation already present in a population of organisms. Other evolutionary forces outside of mutation and natural selection can also play a role or be incorporated into experimental evolution studies, such as genetic drift and gene flow. The organism used is decided by the experimenter, based on whether the hypothesis to be tested involves adaptation through mutation or allele frequency change. A large number of generations are required for adaptive mutation to occur, and experimental evolution via mutation is carried out in viruses or unicellular organisms with rapid generation times, such as bacteria and asexual clonal yeast. Polymorphic populations of asexual or sexual yeast, and multicellular eukaryotes like Drosophila, can adapt to new environments through allele frequency change in standing genetic variation. Organisms with longer generations times, although costly, can be used in experimental evolution. Laboratory studies with foxes and with rodents have shown that notable adaptations can occur within as few as 10–20 generations and experiments with wild guppies have observed adaptations within comparable numbers of generations. More recently, experimentally evolved individuals or populations are often analyzed using whole genome sequencing, an approach known as Evolve and Resequence (E&R). E&R can identify mutations that lead to adaptation in clonal individuals or identify alleles that changed in frequency in polymorphic populations, by comparing the sequences of individuals/populations before and after adaptation. The sequence data makes it possible to pinpoint the site in a DNA sequence that a mutation/allele frequency change occurred to bring about adaptation. The nature of the adaptation and functional follow up studies can shed insight into what effect the mutation/allele has on phenotype.

Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes that are passed on from parent to offspring during reproduction. Different characteristics tend to exist within any given population as a result of mutation, genetic recombination and other sources of genetic variation. Evolution occurs when evolutionary processes such as natural selection and genetic drift act on this variation, resulting in certain characteristics becoming more common or rare within a population. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms and molecules.
In population biology and demography, the generation time is the average time between two consecutive generations in the lineages of a population. In human populations, the generation time typically ranges from 22 to 33 years. Historians sometimes use this to date events, by converting generations into years to obtain rough estimates of time.
Contents
- Experimental studies, by category
- Laboratory phylogenetics
- Epistasis
- Experimental adaptation
- Impact of sex/coinfection
- Muller’s ratchet
- Prisoner’s dilemma
- Coevolution
- References
- Bibliography
- Laboratory phylogenetics 2
- Epistasis 2
- Experimental adaptation 2
- Impact of sex/coinfection 2
- Muller’s ratchet 2
- Prisoner’s dilemma 2
- Coevolution 2