Related Research Articles

Magnetic resonance imaging (MRI) is a medical imaging technique used in radiology to form pictures of the anatomy and the physiological processes of the body. MRI scanners use strong magnetic fields, magnetic field gradients, and radio waves to generate images of the organs in the body. MRI does not involve X-rays or the use of ionizing radiation, which distinguishes it from CT and PET scans. MRI is a medical application of nuclear magnetic resonance (NMR) which can also be used for imaging in other NMR applications, such as NMR spectroscopy.
Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution.
Iterative reconstruction refers to iterative algorithms used to reconstruct 2D and 3D images in certain imaging techniques. For example, in computed tomography an image must be reconstructed from projections of an object. Here, iterative reconstruction techniques are usually a better, but computationally more expensive alternative to the common filtered back projection (FBP) method, which directly calculates the image in a single reconstruction step. In recent research works, scientists have shown that extremely fast computations and massive parallelism is possible for iterative reconstruction, which makes iterative reconstruction practical for commercialization.

In computational modelling, multiphysics simulation is defined as the simultaneous simulation of different aspects of a physical system or systems and the interactions among them. For example, simultaneous simulation of the physical stress on an object, the temperature distribution of the object and the thermal expansion which leads to the variation of the stress and temperature distributions would be considered a multiphysics simulation. Multiphysics simulation is related to multiscale simulation, which is the simultaneous simulation of a single process on either multiple time or distance scales.
Blood-oxygen-level-dependent imaging, or BOLD-contrast imaging, is a method used in functional magnetic resonance imaging (fMRI) to observe different areas of the brain or other organs, which are found to be active at any given time.
Magnetic resonance elastography (MRE) is a form of elastography that specifically leverages MRI to quantify and subsequently map the mechanical properties of soft tissue. First developed and described at Mayo Clinic by Muthupillai et al. in 1995, MRE has emerged as a powerful, non-invasive diagnostic tool, namely as an alternative to biopsy and serum tests for staging liver fibrosis.
In computational chemistry, a constraint algorithm is a method for satisfying the Newtonian motion of a rigid body which consists of mass points. A restraint algorithm is used to ensure that the distance between mass points is maintained. The general steps involved are: (i) choose novel unconstrained coordinates, (ii) introduce explicit constraint forces, (iii) minimize constraint forces implicitly by the technique of Lagrange multipliers or projection methods.
Signal enhancement by extravascular water protons, or SEEP, is a contrast mechanism for functional magnetic resonance imaging (fMRI), which is an alternative to the more commonly employed BOLD contrast. This mechanism for image contrast changes corresponding to changes in neuronal activity was first proposed by Dr. Patrick Stroman in 2001. SEEP contrast is based on changes in tissue water content which arise from the increased production of extracellular fluid and swelling of neurons and glial cells at sites of neuronal activity. Because the dominant sources of MRI signal in biological tissues are water and lipids, an increase in tissue water content is reflected by a local increase in MR signal intensity. A correspondence between BOLD and SEEP signal changes, and sites of activity, has been observed in the brain and appears to arise from the common dependence on changes in local blood flow to cause a change in blood oxygenation or to produce extracellular fluid. The advantage of SEEP contrast is that it can be detected with MR imaging methods which are relatively insensitive to magnetic susceptibility differences between air, tissues, blood, and bone. Such susceptibility differences can give rise to spatial image distortions and areas of low signal, and magnetic susceptibility changes in blood give rise to the BOLD contrast for fMRI. The primary application of SEEP to date has been fMRI of the spinal cord because the bone/tissue interfaces around the spinal cord cause poor image quality with conventional fMRI methods. The disadvantages of SEEP compared to BOLD contrast are that it reveals more localized areas of activity, and in the brain the signal intensity changes are typically lower, and it can therefore be more difficult to detect.
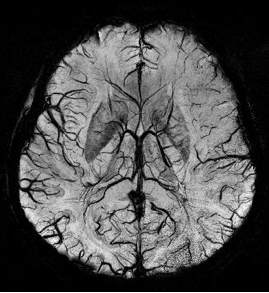
Susceptibility weighted imaging (SWI), originally called BOLD venographic imaging, is an MRI sequence that is exquisitely sensitive to venous blood, hemorrhage and iron storage. SWI uses a fully flow compensated, long echo, gradient recalled echo (GRE) pulse sequence to acquire images. This method exploits the susceptibility differences between tissues and uses the phase image to detect these differences. The magnitude and phase data are combined to produce an enhanced contrast magnitude image. The imaging of venous blood with SWI is a blood-oxygen-level dependent (BOLD) technique which is why it was referred to as BOLD venography. Due to its sensitivity to venous blood SWI is commonly used in traumatic brain injuries (TBI) and for high resolution brain venographies but has many other clinical applications. SWI is offered as a clinical package by Philips and Siemens but can be run on any manufacturer’s machine at field strengths of 1.0 T, 1.5 T, 3.0 T and higher.

Real-time magnetic resonance imaging (RT-MRI) refers to the continuous monitoring ("filming") of moving objects in real time. Because MRI is based on time-consuming scanning of k-space, real-time MRI was possible only with low image quality or low temporal resolution. Using an iterative reconstruction algorithm these limitations have recently been removed: a new method for real-time MRI achieves a temporal resolution of 20 to 30 milliseconds for images with an in-plane resolution of 1.5 to 2.0 mm. Real-time MRI promises to add important information about diseases of the joints and the heart. In many cases MRI examinations may become easier and more comfortable for patients.

Quantitative Susceptibility Mapping (QSM) provides a novel contrast mechanism in Magnetic Resonance Imaging (MRI) different from traditional Susceptibility Weighted Imaging. The voxel intensity in QSM is linearly proportional to the underlying tissue apparent magnetic susceptibility, which is useful for chemical identification and quantification of specific biomarkers including iron, calcium, gadolinium, and super paramagnetic iron oxide (SPIO) nano-particles. QSM utilizes phase images, solves the magnetic field to susceptibility source inverse problem, and generates a three-dimensional susceptibility distribution. Due to its quantitative nature and sensitivity to certain kinds of material, potential QSM applications include standardized quantitative stratification of cerebral microbleeds and neurodegenerative disease, accurate gadolinium quantification in contrast enhanced MRI, and direct monitoring of targeted theranostic drug biodistribution in nanomedicine.

Magnetic resonance imaging of the brain uses magnetic resonance imaging (MRI) to produce high quality two-dimensional or three-dimensional images of the brain and brainstem as well as the cerebellum without the use of ionizing radiation (X-rays) or radioactive tracers.

Philip Batchelor, was a Swiss-British academic in the fields of mathematics and medical imaging.
Christopher J. Hardy is an American physicist and inventor of several magnetic resonance imaging (MRI) subsystem technologies for use in real time MRI and cardiac MR imaging and spectroscopy.
Harmonic phase (HARP) algorithm is a medical image analysis technique capable of extracting and processing motion information from tagged magnetic resonance image (MRI) sequences. It was initially developed by N. F. Osman and J. L. Prince at the Image Analysis and Communications Laboratory at Johns Hopkins University. The method uses spectral peaks in the Fourier domain of tagged MRI, calculating the phase images of their inverse Fourier transforms, which are called harmonic phase (HARP) images. The motion of material points through time is then tracked, under the assumption that the HARP value of a fixed material point is time-invariant. The method is fast and accurate, and has been accepted as one of the most popular tagged MRI analysis methods in medical image processing.
The following is a timeline of numerical analysis after 1945, and deals with developments after the invention of the modern electronic computer, which began during Second World War. For a fuller history of the subject before this period, see timeline and history of mathematics.

Jürgen Klaus Hennig is a German chemist and medical physicist. Internationally he is considered to be one of the pioneers of Magnetic Resonance Imaging for clinical diagnostics. He is the Scientific Director of the Department of Diagnostic Radiology and Chairman of the Magnetic Resonance Development and Application Center (MRDAC) at the University Medical Center Freiburg. In the year 2003 he was awarded the Max Planck Research Award in the category of Biosciences and Medicine.
Synthetic MRI is a simulation method in Magnetic Resonance Imaging (MRI), for generating contrast weighted images based on measurement of tissue properties. The synthetic (simulated) images are generated after an MR study, from parametric maps of tissue properties. It is thereby possible to generate several contrast weightings from the same acquisition. This is different from conventional MRI, where the signal acquired from the tissue is used to generate an image directly, often generating only one contrast weighting per acquisition. The synthetic images are similar in appearance to those normally acquired with an MRI scanner.

JCMsuite is a finite element analysis software package for the simulation and analysis of electromagnetic waves, elasticity and heat conduction. It also allows a mutual coupling between its optical, heat conduction and continuum mechanics solvers. The software is mainly applied for the analysis and optimization of nanooptical and microoptical systems. Its applications in research and development projects include dimensional metrology systems, photolithographic systems, photonic crystal fibers, VCSELs, Quantum-Dot emitters, light trapping in solar cells, and plasmonic systems. The design tasks can be embedded into the high-level scripting languages MATLAB and Python, enabling a scripting of design setups in order to define parameter dependent problems or to run parameter scans.
The history of magnetic resonance imaging (MRI) includes the work of many researchers who contributed to the discovery of nuclear magnetic resonance (NMR) and described the underlying physics of magnetic resonance imaging, starting early in the twentieth century. MR imaging was invented by Paul C. Lauterbur who developed a mechanism to encode spatial information into an NMR signal using magnetic field gradients in September 1971; he published the theory behind it in March 1973. The factors leading to image contrast had been described nearly 20 years earlier by physician and scientist Erik Odeblad and Gunnar Lindström. Among many other researchers in the late 1970s and 1980s, Peter Mansfield further refined the techniques used in MR image acquisition and processing, and in 2003 he and Lauterbur were awarded the Nobel Prize in Physiology or Medicine for their contributions to the development of MRI. The first clinical MRI scanners were installed in the early 1980s and significant development of the technology followed in the decades since, leading to its widespread use in medicine today.
References
- Stöcker T, Vahedipour K, Shah NJ (2008). "HPC Simulation of Magnetic Resonance Imaging". Adv Par Comp. 15 (1): 155–164.
- Vahedipour, K; Stöcker T; Shah NJ (2008). "jemris 2.0: High Performance Computing Simulator for MRI". Proceedings of the 26th ESMRMB. Valencia, ES.
- Vahedipour, K; Stöcker T; Shah NJ (2009). "Fast MR Simulations with jemris 2.1 - Disclosing the Secrets of MRI Sequence Development". Proceedings of the 17th ISMRM. Honolulu, HI, USA.
- Stöcker T, Vahedipour K, Shah NJ (2010). "High Performance Computing MRI Simulations". Magn Reson Med. 64 (1): 186–93. doi:10.1002/mrm.22406. PMID 20577987. S2CID 21797770.
- Nieuwstadt HA; et al. (2014). "Numerical simulations of carotid MRI quantify the accuracy in measuring atherosclerotic plaque components in vivo". Magn Reson Med. 72 (1): 188–201. doi:10.1002/mrm.24905. PMID 23943090. S2CID 22939029.