In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the basis for biological inheritance. The cell possesses the distinctive property of division, which makes replication of DNA not essential.
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950 when she noticed that streaks of mixtures of two E. coli strains, one of which treated with ultraviolet light, was "nibbled and plaqued". The wild type of this virus has a temperate lifecycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase ; mutant strains are unable to lysogenize cells – instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the Caulimoviridae family, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.

The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that only use the lytic cycle are called virulent phages.
This is a list of topics in molecular biology. See also index of biochemistry articles.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for P trait of P element and it is found only in wild flies.
In molecular biology and genetics, the sense of nucleic acid molecules is the nature of their roles and their complementary molecules' nucleic acid units' roles in specifying amino acids. Depending on the context within molecular biology, sense may have slightly different meanings.
fis is an E. coli gene encoding the Fis protein. The regulation of this gene is more complex than most other genes in the E. coli genome, as Fis is an important protein which regulates expression of other genes. It is supposed that fis is regulated by H-NS, IHF and CRP. It also regulates its own expression (autoregulation). Fis is one of the most abundant DNA binding proteins in Escherichia coli under nutrient-rich growth conditions.
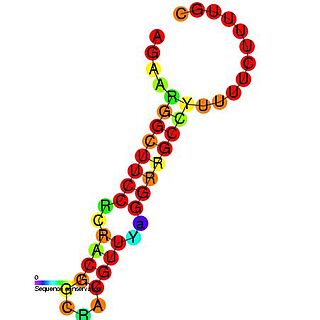
In molecular biology ctRNA is a plasmid encoded noncoding RNA that binds to the mRNA of repB and causes translational inhibition. ctRNA is encoded by plasmids and functions in rolling circle replication to maintain a low copy number. In Corynebacterium glutamicum, it achieves this by antisense pairing with the mRNA of RepB, a replication initiation protein. In Enterococcus faecium the plasmid pJB01 contains three open reading frames, copA, repB, and repC. The pJB01 ctRNA is coded on the opposite strand from the copA/repB intergenic region and partially overlaps an atypical ribosome binding site for repB.
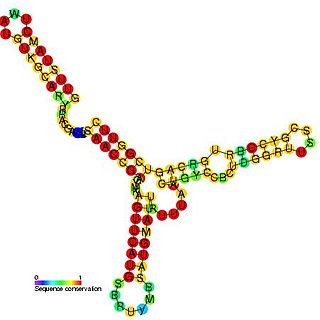
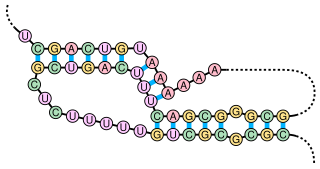
Anti-Q RNA is a small ncRNA from the conjugal plasmid pCF10 of Enterococcus faecalis. It is coded in cis to its regulatory target, prgQ, but can also act in trans. Anti-Q is known to interact with nascent prgQ transcripts to allow formation of an intrinsic terminator, or attenuator, thus preventing transcription of downstream genes. This mode of regulation is essentially the same as that of the countertranscript-driven attenuators that control copy number in pT181, pAMbeta1 and pIP501 and related Staphylococcal plasmids.
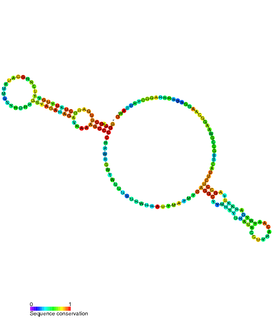
The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.

A circular prokaryote chromosome is a chromosome in bacteria and archaea, in the form of a molecule of circular DNA. Unlike the linear DNA of most eukaryotes, typical prokaryote chromosomes are circular.
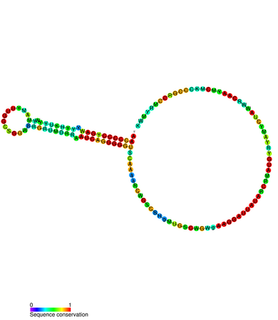
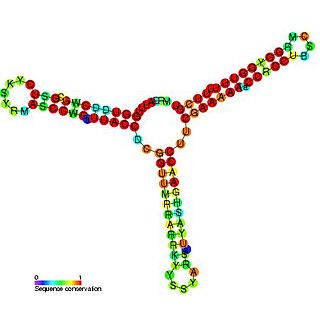
In plasmids, the regulatory region of repBA gene forms a pseudoknot. The repA gene, which encodes a protein likely to function as an initiator for replication, and the repB gene are translationally coupled. The leader sequence of the repA mRNA contains two complementary sequences of 8 bases. Base-pairing between these two sequences forms a pseudoknot which is essential for translation. The first of these complementary sequences is found within a stem-loop, which forms a target for RNAI. Binding of RNAI to this stem-loop inhibits pseudoknot formation and translation of RepA.
The gua operon is responsible for regulating the synthesis of guanosine mono phosphate(GMP), a purine nucleotide, from inosine monophosphate. It consists of two structural genes guaB(encodes for IMP dehydrogenase or IMPDH) and guaA(encodes for GMP synthetase) apart from the promoter and operator region.