| Apocytochr_F_C | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 cytochrome f from the b6f complex of Phormidium laminosum | |||||||||
| Identifiers | |||||||||
| Symbol | Apocytochr_F_C | ||||||||
| Pfam | PF01333 | ||||||||
| Pfam clan | CL0105 | ||||||||
| InterPro | IPR002325 | ||||||||
| PROSITE | PDOC00169 | ||||||||
| SCOP | 1ctm | ||||||||
| SUPERFAMILY | 1ctm | ||||||||
| TCDB | 3.D.3 | ||||||||
| OPM superfamily | 92 | ||||||||
| OPM protein | 3h1j | ||||||||
| |||||||||
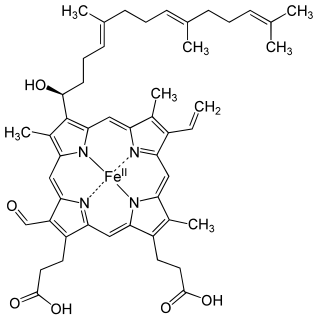
Cytochrome f is the largest subunit of cytochrome b6f complex (plastoquinol—plastocyanin reductase; EC 1.10.99.1). In its structure and functions, the cytochrome b6f complex bears extensive analogy to the cytochrome bc1 complex of mitochondria and photosynthetic purple bacteria. Cytochrome f (cyt f) plays a role analogous to that of cytochrome c1, in spite of their different structures. [1]

The cytochrome b6f complex is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, that catalyzes the transfer of electrons from plastoquinol to plastocyanin. The reaction is analogous to the reaction catalyzed by cytochrome bc1 of the mitochondrial electron transport chain. During photosynthesis, the cytochrome b6f complex is one step along the chain that transfers electrons from Photosystem II to Photosystem I, and at the same time pumps protons into the thylakoid space that contribute to create an electrochemical (energy) gradient which is later used to synthesize ATP from ADP.
The Enzyme Commission number is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the respective enzyme.
Contents
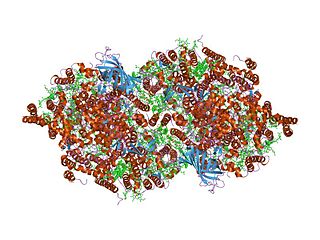
The 3D structure of Brassica rapa (Turnip) cyt f has been determined. [2] The lumen-side segment of cyt f includes two structural domains: a small one above a larger one that, in turn, is on top of the attachment to the membrane domain. The large domain consists of an anti-parallel beta-sandwich and a short haem-binding peptide, which form a three-layer structure. The small domain is inserted between beta-strands F and G of the large domain and is an all-beta domain. The haem nestles between two short helices at the N terminus of cyt f. Within the second helix is the sequence motif for the c-type cytochromes, CxxCH (residues 21-25), which is covalently attached to the haem through thioether bonds to Cys-21 and Cys-24. His-25 is the fifth haem iron ligand. The sixth haem iron ligand is the alpha-amino group of Tyr-1 in the first helix. [2] Cyt f has an internal network of water molecules that may function as a proton wire. [2] The water chain appears to be a conserved feature of cyt f.

The turnip or white turnip is a root vegetable commonly grown in temperate climates worldwide for its white, fleshy taproot. The word turnip is a compound of tur- as in turned/rounded on a lathe and neep, derived from Latin napus, the word for the plant. Small, tender varieties are grown for human consumption, while larger varieties are grown as feed for livestock. In the north of England, Scotland, Ireland, Cornwall and eastern Canada (Newfoundland), turnip often refers to rutabaga, a larger, yellow root vegetable in the same genus (Brassica) also known as swede.

Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a residue indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, and dual polarisation interferometry to determine the structure of proteins.

The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group donates a hydrogen bond to the backbone C=O group of the amino acid located three or four residues earlier along the protein sequence.