| Drosophila roX1 ncRNA | |
|---|---|
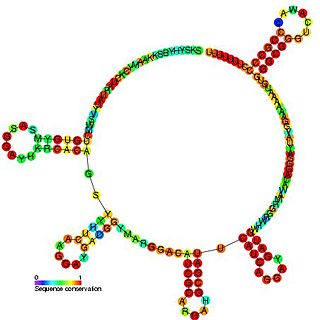
Conserved secondary structure of roX1 RNA | |
| Identifiers | |
| Symbol | roX1 |
| Rfam | RF01667 |
| Other data | |
| RNA type | Cis-regulatory element |
| Domain(s) | Drosophila |
RoX RNA is a non-coding RNA (ncRNA) present in the male-specific lethal (MSL) complex and is required for sex dosage compensation in Drosophila . As males only contain one X chromosome, male flies dosage compensate for the X chromosome by hyper-transcribing the X chromosome. This is achieved by the MSL complex binding to the X chromosome and inducing histone H4 lysine 16 acetylation and allows for the formation of euchromatin. [1] [2] These ncRNAs were first discovered in RNA extracted from neuronal cells. [3] [4]

A non-coding RNA (ncRNA) is an RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Dosage compensation is the process by which organisms equalize the expression of genes between members of different biological sexes. Across species, different sexes are often characterized by different types and numbers of sex chromosomes. In order to neutralize the large difference in gene dosage produced by differing numbers of sex chromosomes among the sexes, various evolutionary branches have acquired various methods to equalize gene expression among the sexes. Because sex chromosomes contain different numbers of genes, different species of organisms have developed different mechanisms to cope with this inequality. Replicating the actual gene is impossible; thus organisms instead equalize the expression from each gene. For example, in humans, females (XX) silence the transcription of one X chromosome of each pair, and transcribe all information from the other, expressed X chromosome. Thus, human females have the same number of expressed X-linked genes as do human males (XY), both sexes having essentially one X chromosome per cell, from which to transcribe and express genes.

Drosophila is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies ; tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of Drosophila in particular, D. melanogaster, has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "Drosophila" are often used synonymously with D. melanogaster in modern biological literature. The entire genus, however, contains more than 1,500 species and is very diverse in appearance, behavior, and breeding habitat.
Contents
Two roX RNAs have been identified in the MSL complex from Drosophila melanogaster and have been shown to be conserved across different drosophila species. The two roX RNAs, known as roX1 and roX2 have been shown to differ in primary sequence and sequence length but despite these differences they both contain a MSL binding site and are functionally redundant. [5] [6] Cloning roX genes and subsequent sequence alignment from a number of different drosophila species identified several conserved regions and it was also noticed that roX RNA is only present in male flies. [7]

Drosophila melanogaster is a species of fly in the family Drosophilidae. The species is known generally as the common fruit fly or vinegar fly. Starting with Charles W. Woodworth's proposal of the use of this species as a model organism, D. melanogaster continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and life history evolution. As of 2017, eight Nobel prizes had been awarded for research using Drosophila.

Cloning is the process of producing genetically identical individuals of an organism either naturally or artificially. In nature, many organisms produce clones through asexual reproduction. Cloning in biotechnology refers to the process of creating clones of organisms or copies of cells or DNA fragments. Beyond biology, the term refers to the production of multiple copies of digital media or software.

In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the edit distance cost between strings in a natural language or in financial data.
Secondary structure predictions predicted a stem loop to be present at the 3' end of the roX RNA transcripts. Mutagenesis within this region that either altered the sequence or prevented stem loop formation was shown to be lethal in male flies as they were unable to dosage compensate as these mutations prevented MSL binding to the X chromosome. It was also shown that in the presence of mutant roX RNA the MSL complex was unable to localize on the X chromosome and mislocalize to the heterochromatic chromocenter. This suggests that roX RNA plays a role in directing MSL complex to the X chromosome. [7] Sequence alignment of roX2 identified several regions of sequence conservation known as Gub regions and are absent in roX1. Many of these Gub regions are thought to be redundant as mutating has no effect however, Gub1 which is present within the 3' stem loop region is thought to be a key motif required for roX2 as Gub1 mutations were shown to be lethal. These studies show that roX RNA are essential for dosage compensation where that the 3' stem loop region in the roX RNA are crucial for MSL localization and it is thought to be the motif that allows the two roX RNAs to be functionally redundant.
Mutagenesis is a process by which the genetic information of an organism is changed, resulting in a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures. In nature mutagenesis can lead to cancer and various heritable diseases, but it is also a driving force of evolution. Mutagenesis as a science was developed based on work done by Hermann Muller, Charlotte Auerbach and J. M. Robson in the first half of the 20th century.

In biology and especially genetics, a mutant is an organism or a new genetic character arising or resulting from an instance of mutation, which is generally an alteration of the DNA sequence of the genome or chromosome of an organism. The term mutant is also applied to a virus with an alteration in its nucleotide sequence whose genome is RNA, rather than DNA. In multicellular eukaryotes, a DNA sequence may be altered in an individual somatic cell that then gives rise to a mutant somatic cell lineage as happens in cancer progression. Also in eukaryotes, alteration of a mitochondrial or plastid DNA sequence may give rise to a mutant lineage that is inherited separately from mutant genotypes in the nuclear genome. The natural occurrence of genetic mutations is integral to the process of evolution. The study of mutants is an integral part of biology; by understanding the effect that a mutation in a gene has, it is possible to establish the normal function of that gene.