Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.

The lactose operon is an operon required for the transport and metabolism of lactose in E. coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
The galactose permease or GalP found in Escherichia coli is an integral membrane protein involved in the transport of monosaccharides, primarily hexoses, for utilization by E. coli in glycolysis and other metabolic and catabolic pathways (3,4). It is a member of the Major Facilitator Super Family (MFS) and is homologue of the human GLUT1 transporter (4). Below you will find descriptions of the structure, specificity, effects on homeostasis, expression, and regulation of GalP along with examples of several of its homologues.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

A colicin is a type of bacteriocin produced by and toxic to some strains of Escherichia coli. Colicins are released into the environment to reduce competition from other bacterial strains. Colicins bind to outer membrane receptors, using them to translocate to the cytoplasm or cytoplasmic membrane, where they exert their cytotoxic effect, including depolarisation of the cytoplasmic membrane, DNase activity, RNase activity, or inhibition of murein synthesis.
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.

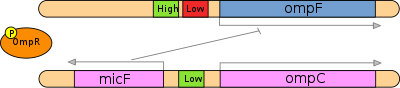
The MicC non-coding RNA is located between the ompN and ydbK genes in E. coli. This Hfq-associated RNA is thought to be a regulator of the expression level of the OmpC porin protein, with a 5′ region of 22 nucleotides potentially forming an antisense interaction with the ompC mRNA. Along with MicF RNA this family may act in conjunction with EnvZ-OmpR two-component system to control the OmpF/OmpC protein ratio in response to a variety of environmental stimuli. The expression of micC was shown to be increased in the presence of beta-lactam antibiotics.

The micF RNA is a non-coding RNA stress response gene found in Escherichia coli and related bacteria that post-transcriptionally controls expression of the outer membrane porin gene ompF. The micF gene encodes a non-translated 93 nucleotide antisense RNA that binds its target ompF mRNA and regulates ompF expression by inhibiting translation and inducing degradation of the message. In addition, other factors, such as the RNA chaperone protein StpA also play a role in this regulatory system. The expression of micF is controlled by both environmental and internal stress factors. Four transcriptional regulators are known to bind the micF promoter region and activate micF expression.

General bacterial porins are a family of porin proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

In the field of molecular biology, a two-component regulatory system serves as a basic stimulus-response coupling mechanism to allow organisms to sense and respond to changes in many different environmental conditions. Two-component systems typically consist of a membrane-bound histidine kinase that senses a specific environmental stimulus and a corresponding response regulator that mediates the cellular response, mostly through differential expression of target genes. Although two-component signaling systems are found in all domains of life, they are most common by far in bacteria, particularly in Gram-negative and cyanobacteria; both histidine kinases and response regulators are among the largest gene families in bacteria. They are much less common in archaea and eukaryotes; although they do appear in yeasts, filamentous fungi, and slime molds, and are common in plants, two-component systems have been described as "conspicuously absent" from animals.

Histidine kinases (HK) are multifunctional, and in non-animal kingdoms, typically transmembrane, proteins of the transferase class of enzymes that play a role in signal transduction across the cellular membrane. The vast majority of HKs are homodimers that exhibit autokinase, phosphotransfer, and phosphatase activity. HKs can act as cellular receptors for signaling molecules in a way analogous to tyrosine kinase receptors (RTK). Multifunctional receptor molecules such as HKs and RTKs typically have portions on the outside of the cell that bind to hormone- or growth factor-like molecules, portions that span the cell membrane, and portions within the cell that contain the enzymatic activity. In addition to kinase activity, the intracellular domains typically have regions that bind to a secondary effector molecule or complex of molecules that further propagate signal transduction within the cell. Distinct from other classes of protein kinases, HKs are usually parts of a two-component signal transduction mechanisms in which HK transfers a phosphate group from ATP to a histidine residue within the kinase, and then to an aspartate residue on the receiver domain of a response regulator protein. More recently, the widespread existence of protein histidine phosphorylation distinct from that of two-component histidine kinases has been recognised in human cells. In marked contrast to Ser, Thr and Tyr phosphorylation, the analysis of phosphorylated Histidine using standard biochemical and mass spectrometric approaches is much more challenging, and special procedures and separation techniques are required for their preservation alongside classical Ser, Thr and Tyr phosphorylation on proteins isolated from human cells.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviateion for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.
The capsule biosynthesis, or CPS operon, is a section of the genome present in some Escherichia coli, of which regulates the production of polysaccharides making up the bacterial capsule. These polysaccharides help protect the bacteria from harsh environments, toxic chemicals, and bacteriophages.

A response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.

The Phosphate (Pho) regulon is a regulatory mechanism used for the conservation and management of inorganic phosphate within the cell. It was first discovered in Escherichia coli as an operating system for the bacterial strain, and was later identified in other species. The Pho system is composed of various components including extracellular enzymes and transporters that are capable of phosphate assimilation in addition to extracting inorganic phosphate from organic sources. This is an essential process since phosphate plays an important role in cellular membranes, genetic expression, and metabolism within the cell. Under low nutrient availability, the Pho regulon helps the cell survive and thrive despite a depletion of phosphate within the environment. When this occurs, phosphate starvation-inducible (psi) genes activate other proteins that aid in the transport of inorganic phosphate.