Transmembrane domain usually denotes a transmembrane segment of single alpha helix of a transmembrane protein. More broadly, a transmembrane domain is any membrane-spanning protein domain.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane to which it is permanently attached. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane.
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP or NAD+ as cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, chloroplasts and mitochondria, including respiratory complexes I, II and III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix.

The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor.
Scleroproteins or fibrous proteins constitute one of the three main types of proteins. There are many scleroprotein superfamilies including keratin, collagen, elastin, and fibroin. The roles of such proteins include protection and support, forming connective tissue, tendons, bone matrices, and muscle fiber.
A coiled coil is a structural motif in proteins in which 2–7 alpha-helices are coiled together like the strands of a rope. Many coiled coil-type proteins are involved in important biological functions such as the regulation of gene expression, e.g. transcription factors. Notable examples are the oncoproteins c-Fos and c-jun, as well as the muscle protein tropomyosin.

SNARE proteins — "SNAPREceptor" — are a large protein complex consisting of at least 24 members in yeasts and more than 60 members in mammalian cells. The primary role of SNARE proteins is to mediate vesicle fusion, that is, the fusion of vesicles with their target membrane bound compartments. The best studied SNAREs are those that mediate docking of synaptic vesicles with the presynaptic membrane in neurons. These SNAREs are the targets of the bacterial neurotoxins responsible for botulism and tetanus.

A basic helix-loop-helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
Knobs into holes packing is a protein folding motif that occurs mainly in alpha helix or coiled coil domains. One such example is fibrinogen fibril formation.
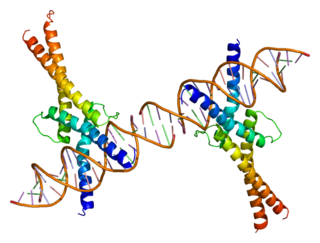
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

Sterol regulatory element-binding protein 2 (SREBP-2) also known as sterol regulatory element binding transcription factor 2 (SREBF2) is a protein that in humans is encoded by the SREBF2 gene.
Orientations of Proteins in Membranes (OPM) database provides spatial positions of membrane protein structures with respect to the lipid bilayer. Positions of the proteins are calculated using an implicit solvation model of the lipid bilayer. The results of calculations were verified against experimental studies of spatial arrangement of transmembrane and peripheral proteins in membranes.
Bacteriophage f1 is structurally classified as a class I filamentous phage, and is closely related to the other Ff phages, such as M13 and phage fd.

The Protein Property Prediction and Testing Database (PPT-DB) is a collection of protein property databases for over 20 different protein properties including secondary structure, trans-membrane helices and beta barrels, accessible surface area, signal peptides, and more. PPT-DB is published in 2008. Its protocol is available via PPT-DB website

AmphipathicLipid Packing Sensor (ALPS) motifs were first identified in 2005 in ARFGAP1 and have been reviewed.
Membranome database provides structural and functional information about more than 6000 single-pass (bitopic) transmembrane proteins from Homo sapiens, Arabidopsis thaliana, Dictyostelium discoideum, Saccharomyces cerevisiae, Escherichia coli and Methanocaldococcus jannaschii. Bitopic membrane proteins consist of a single transmembrane alpha-helix connecting water-soluble domains of the protein situated at the opposite sides of a biological membrane. These proteins are frequently involved in the signal transduction and communication between cells in multicellular organisms.

Bitopic proteins are transmembrane proteins that span the lipid bilayer only one time. These proteins may constitute up to 50% of all transmembrane proteins, depending on the organism, and contribute significantly to the network of interactions between different proteins in cells, including interactions via transmembrane helices. They usually include one or several water-soluble domains situated at the different sides of biological membranes, for example in single-pass transmembrane receptors. Some of them are small and serve as regulatory or structure-stabilizing subunits in large multi-protein transmembrane complexes, such as photosystems or the respiratory chain.










