
Pyrrolysine is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group, a carboxylic acid group. Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH.
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences.
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.

Aconitase is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
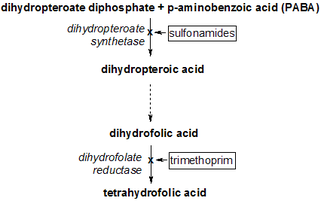
Dihydropteroate synthase is an enzyme classified under EC 2.5.1.15. It produces dihydropteroate in bacteria, but it is not expressed in most eukaryotes including humans. This makes it a useful target for sulfonamide antibiotics, which compete with the PABA precursor.

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
Biotin synthase (BioB) is an enzyme that catalyzes the conversion of dethiobiotin (DTB) to biotin; this is the final step in the biotin biosynthetic pathway. Biotin, also known as vitamin B7, is a cofactor used in carboxylation, decarboxylation, and transcarboxylation reactions in many organisms including humans. Biotin synthase is an S-Adenosylmethionine (SAM) dependent enzyme that employs a radical mechanism to thiolate dethiobiotin, thus converting it to biotin.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
(Methyl-Co methanol-specific corrinoid protein):coenzyme M methyltransferase is an enzyme with systematic name methylated methanol-specific corrinoid protein:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction
(Methyl-Co methylamine-specific corrinoid protein):coenzyme M methyltransferase is an enzyme with systematic name methylated monomethylamine-specific corrinoid protein:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction
Methylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name monomethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
Dimethylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name dimethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
Trimethylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name trimethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
Methylated-thiol-coenzyme M methyltransferase is an enzyme with systematic name methylated-thiol:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction:
Tetramethylammonium-corrinoid protein Co-methyltransferase is an enzyme with systematic name tetramethylammonium:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
(Methyl-Co tetramethylammonium-specific corrinoid protein):coenzyme M methyltransferase is an enzyme with systematic name methylated tetramethylammonium-specific corrinoid protein:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction
5-methyltetrahydrofolate:corrinoid/iron-sulfur protein Co-methyltransferase is an enzyme with systematic name 5-methyltetrahydrofolate:corrinoid/iron-sulfur protein methyltransferase. This enzyme catalyses the following chemical reaction

Acetyl-CoA synthase (ACS), not to be confused with Acetyl-CoA synthetase or Acetate-CoA ligase, is a nickel-containing enzyme involved in the metabolic processes of cells. Together with Carbon monoxide dehydrogenase (CODH), it forms the bifunctional enzyme Acetyl-CoA Synthase/Carbon Monoxide Dehydrogenase (ACS/CODH) found in anaerobic organisms such as archaea and bacteria. The ACS/CODH enzyme works primarily through the Wood–Ljungdahl pathway which converts carbon dioxide to Acetyl-CoA. The recommended name for this enzyme is CO-methylating acetyl-CoA synthase.
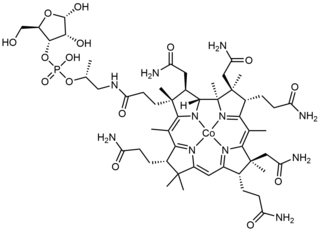
Cobamide is a naturally occurring chemical compound containing cobalt in the corrinoid family of macrocyclic complexes. Cobamide works as a coenzyme with some enzymes in bacteria. The cobalt atom may have a transferable methyl group attached. It is used for example in 5-methyltetrahydrosarcinapterin:corrinoid/iron-sulfur protein Co-methyltransferase.









