Related Research Articles

Staphylococcus aureus is a Gram-positive, round-shaped bacterium a member of the Firmicutes, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. The emergence of antibiotic-resistant strains of S. aureus such as methicillin-resistant S. aureus (MRSA) is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Neomycin is an aminoglycoside antibiotic that displays bactericidal activity against gram-negative aerobic bacilli and some anaerobic bacilli where resistance has not yet arisen. It is generally not effective against gram-positive bacilli and anaerobic gram-negative bacilli. Neomycin comes in oral and topical formulations, including creams, ointments, and eyedrops. Neomycin belongs to the aminoglycoside class of antibiotics that contain two or more amino sugars connected by glycosidic bonds.

Puromycin is an antibiotic protein synthesis inhibitor which causes premature chain termination during translation.
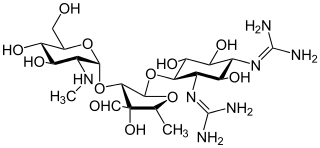
Aminoglycoside is a medicinal and bacteriologic category of traditional Gram-negative antibacterial medications that inhibit protein synthesis and contain as a portion of the molecule an amino-modified glycoside (sugar). The term can also refer more generally to any organic molecule that contains amino sugar substructures. Aminoglycoside antibiotics display bactericidal activity against Gram-negative aerobes and some anaerobic bacilli where resistance has not yet arisen but generally not against Gram-positive and anaerobic Gram-negative bacteria.
Integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes. These genes are embedded in a specific genetic structure called gene cassette that generally carries one promoterless open reading frame (ORF) together with a recombination site (attC). Integron cassettes are incorporated to the attI site of the integron platform by site-specific recombination reactions mediated by the integrase.

Kanamycin A, often referred to simply as kanamycin, is an antibiotic used to treat severe bacterial infections and tuberculosis. It is not a first line treatment. It is used by mouth, injection into a vein, or injection into a muscle. Kanamycin is recommended for short-term use only, usually from 7 to 10 days. As with most antibiotics, it is ineffective in viral infections.

Viomycin is a member of the tuberactinomycin family, a group of nonribosomal peptide antibiotics exhibiting anti-tuberculosis activity. The tuberactinomycin family is an essential component in the drug cocktail currently used to fight infections of Mycobacterium tuberculosis. Viomycin was the first member of the tuberactinomycins to be isolated and identified, and was used to treat TB until it was replaced by the less toxic, but structurally related compound, capreomycin. The tuberactinomycins target bacterial ribosomes, binding RNA and disrupting bacterial protein synthesis and certain forms of RNA splicing. Viomycin is produced by the actinomycete Streptomyces puniceus.

Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin.
A gene cassette is a type of mobile genetic element that contains a gene and a recombination site. Each cassette usually contains a single gene and tends to be very small; on the order of 500–1000 base pairs. They may exist incorporated into an integron or freely as circular DNA. Gene cassettes can move around within an organism's genome or be transferred to another organism in the environment via horizontal gene transfer. These cassettes often carry antibiotic resistance genes. An example would be the kanMX cassette which confers kanamycin resistance upon bacteria.
The rpoB gene encodes the β subunit of bacterial RNA polymerase. rpoB is also found in plant chloroplasts where it forms the beta subunit of the plastid-encoded RNA polymerase (PEP). An inhibitor of transcription in bacteria, tagetitoxin, also inhibits PEP, showing that the complex found in plants is very similar to the homologous enzyme in bacteria. It codes for 1342 amino acids, making it the second-largest polypeptide in the bacterial cell. It is the site of mutations that confer resistance to the rifamycin antibacterial agents, such as rifampin. Mutations in rpoB that confer resistance to rifamycins do so by altering residues of the rifamycin binding site on RNA polymerase, thereby reducing rifamycin binding affinity for rifamycins

Hygromycin B is an antibiotic produced by the bacterium Streptomyces hygroscopicus. It is an aminoglycoside that kills bacteria, fungi and higher eukaryotic cells by inhibiting protein synthesis.

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.
In enzymology, an aminoglycoside N6'-acetyltransferase (EC 2.3.1.82) is an enzyme that catalyzes the chemical reaction

A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.
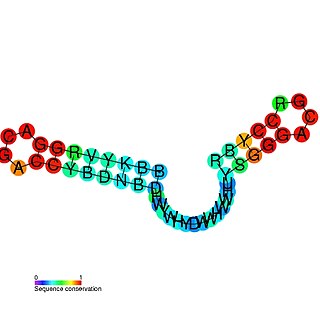
The mini-ykkC RNA motif was discovered as a putative RNA structure that is conserved in bacteria. The motif consists of two conserved stem-loops whose terminal loops contain the RNA sequence ACGR, where R represents either A or G. Mini-ykkC RNAs are widespread in Proteobacteria, but some are predicted in other phyla of bacteria. It was expected that the RNAs are cis-regulatory elements, because they are typically located upstream of protein-coding genes.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. The plasmids can be transferred between bacteria within the same species or between different species via conjugation. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antibiotic usage in human and veterinary medicine.

Neisseria gonorrhoeae, the bacterium that causes the sexually transmitted infection gonorrhea, has developed antibiotic resistance to many antibiotics. The bacteria was first identified in 1879.
16S rRNA (adenine1408-N1)-methyltransferase (EC 2.1.1.180, kanamycin-apramycin resistance methylase, 16S rRNA:m1A1408 methyltransferase, KamB, NpmA, 16S rRNA m1A1408 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (adenine1408-N1)-methyltransferase. This enzyme catalyses the following chemical reaction

Nourseothricin (NTC) is a member of the streptothricin-class of aminoglycoside antibiotics produced by Streptomyces species. Chemically, NTC is a mixture of the related compounds streptothricin C, D, E, and F. NTC inhibits protein synthesis by inducing miscoding. It is used as a selection marker for a wide range of organisms including bacteria, yeast, filamentous fungi, and plant cells. It is not known to have adverse side-effects on positively selected cells, a property cardinal to a selection drug.
References
- 1 2 Jia, Xu; Zhang, Jing; Sun, Wenxia; He, Weizhi; Jiang, Hengyi; Chen, Dongrong; Murchie, Alastair I. H. (2013-01-17). "Riboswitch control of aminoglycoside antibiotic resistance". Cell. 152 (1–2): 68–81. doi: 10.1016/j.cell.2012.12.019 . ISSN 1097-4172. PMID 23332747.
- 1 2 Roth A, Breaker RR (June 2013). "Integron attI1 sites, not riboswitches, associate with antibiotic resistance genes". Cell. 153 (7): 1417–1418. doi:10.1016/j.cell.2013.05.043. PMC 4626886 . PMID 23791167.
- ↑ Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, Murchie AI (June 2013). "Riboswitch regulation of aminoglycoside resistance acetyl and adenyl transferases". Cell. 153 (7): 1419–1420. doi: 10.1016/j.cell.2013.05.050 . PMID 23791168.