Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
The urea cycle (also known as the ornithine cycle) is a cycle of biochemical reactions that produces urea (NH2)2CO from ammonia (NH3). This cycle occurs in ureotelic organisms. The urea cycle converts highly toxic ammonia to urea for excretion. This cycle was the first metabolic cycle to be discovered (Hans Krebs and Kurt Henseleit, 1932), five years before the discovery of the TCA cycle. This cycle was described in more detail later on by Ratner and Cohen. The urea cycle takes place primarily in the liver and, to a lesser extent, in the kidneys.
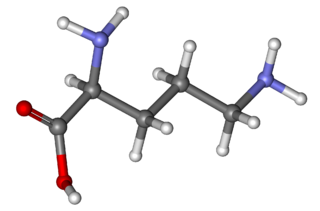
Ornithine is a non-proteinogenic amino acid that plays a role in the urea cycle. Ornithine is abnormally accumulated in the body in ornithine transcarbamylase deficiency. The radical is ornithyl.

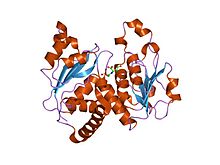
Ornithine transcarbamylase (OTC) is an enzyme that catalyzes the reaction between carbamoyl phosphate (CP) and ornithine (Orn) to form citrulline (Cit) and phosphate (Pi). There are two classes of OTC: anabolic and catabolic. This article focuses on anabolic OTC. Anabolic OTC facilitates the sixth step in the biosynthesis of the amino acid arginine in prokaryotes. In contrast, mammalian OTC plays an essential role in the urea cycle, the purpose of which is to capture toxic ammonia and transform it into urea, a less toxic nitrogen source, for excretion.

Aspartate carbamoyltransferase catalyzes the first step in the pyrimidine biosynthetic pathway.
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined together to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

N-Acetylglutamic acid (also referred to as N-acetylglutamate, abbreviated NAG, chemical formula C7H11NO5) is biosynthesized from glutamate and acetylornithine by ornithine acetyltransferase, and from glutamic acid and acetyl-CoA by the enzyme N-acetylglutamate synthase. The reverse reaction, hydrolysis of the acetyl group, is catalyzed by a specific hydrolase. It is the first intermediate involved in the biosynthesis of arginine in prokaryotes and simple eukaryotes and a regulator in the process known as the urea cycle that converts toxic ammonia to urea for excretion from the body in vertebrates.
Carbamoyl phosphate synthetase I is a ligase enzyme located in the mitochondria involved in the production of urea. Carbamoyl phosphate synthetase I transfers an ammonia molecule from glutamine or glutamate to a molecule of bicarbonate that has been phosphorylated by a molecule of ATP. The resulting carbamate is then phosphorylated with another molecule of ATP. The resulting molecule of carbamoyl phosphate leaves the enzyme.
Pyrimidine biosynthesis occurs both in the body and through organic synthesis.

CAD protein is a trifunctional multi-domain enzyme involved in the first three steps of pyrimidine biosynthesis. De-novo synthesis starts with cytosolic carbamoylphosphate synthetase II which uses glutamine, carbon dioxide and ATP. This enzyme is inhibited by uridine triphosphate.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
Carbamoyl phosphate synthetase II is an enzyme that catalyzes the reactions that produce carbamoyl phosphate in the cytosol. Its systemic name is hydrogen-carbonate:L-glutamine amido-ligase .

In enzymology, an aspartate-semialdehyde dehydrogenase is an enzyme that is very important in the biosynthesis of amino acids in prokaryotes, fungi, and some higher plants. It forms an early branch point in the metabolic pathway forming lysine, methionine, leucine and isoleucine from aspartate. This pathway also produces diaminopimelate which plays an essential role in bacterial cell wall formation. There is particular interest in ASADH as disabling this enzyme proves fatal to the organism giving rise to the possibility of a new class of antibiotics, fungicides, and herbicides aimed at inhibiting it.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction
In enzymology, a N-acetylornithine carbamoyltransferase (EC 2.1.3.9) is an enzyme that catalyzes the chemical reaction

Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, is an enzyme responsible for catalyzing the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

In enzymology, diaminopimelate decarboxylase, also known as diaminopimelic acid decarboxylase, DAPDC, meso-diaminopimelate decarboxylase, DAP-decarboxylase, and meso-2,6-diaminoheptanedioate carboxy-lyase, is an enzyme that catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction
In enzymology, an acetylornithine transaminase (EC 2.6.1.11) is an enzyme that catalyzes the chemical reaction
N-succinylornithine carbamoyltransferase (EC 2.1.3.11, succinylornithine transcarbamylase, N-succinyl-L-ornithine transcarbamylase, SOTCase) is an enzyme with systematic name carbamoyl phosphate:N2-succinyl-L-ornithine carbamoyltransferase. This enzyme catalyses the following chemical reaction