
Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.
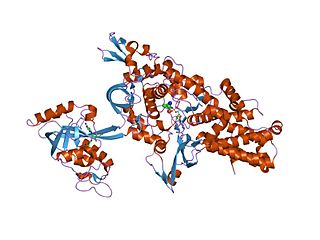
An aminoacyl-tRNA synthetase, also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.
In molecular biology, initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.

The trefoil knot fold is a protein fold in which the protein backbone is twisted into a trefoil knot shape. "Shallow" knots in which the tail of the polypeptide chain only passes through a loop by a few residues are uncommon, but "deep" knots in which many residues are passed through the loop are extremely rare. Deep trefoil knots have been found in the SPOUT superfamily. including methyltransferase proteins involved in posttranscriptional RNA modification in all three domains of life, including bacterium Thermus thermophilus and proteins, in archaea and in eukaryota.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM.

In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.
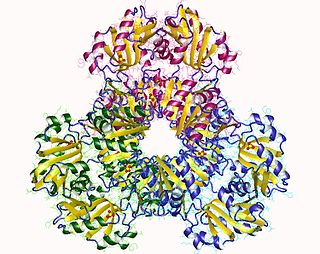
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.

50S is the larger subunit of the 70S ribosome of prokaryotes, i.e. bacteria and archaea. It is the site of inhibition for antibiotics such as macrolides, chloramphenicol, clindamycin, and the pleuromutilins. It includes the 5S ribosomal RNA and 23S ribosomal RNA.

The prokaryotic small ribosomal subunit, or 30S subunit, is the smaller subunit of the 70S ribosome found in prokaryotes. It is a complex of the 16S ribosomal RNA (rRNA) and 19 proteins. This complex is implicated in the binding of transfer RNA to messenger RNA (mRNA). The small subunit is responsible for the binding and the reading of the mRNA during translation. The small subunit, both the rRNA and its proteins, complexes with the large 50S subunit to form the 70S prokaryotic ribosome in prokaryotic cells. This 70S ribosome is then used to translate mRNA into proteins.

In enzymology, a phenylalanine—tRNA ligase is an enzyme that catalyzes the chemical reaction
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction

Phenylalanyl-tRNA synthetase beta chain is an enzyme that in humans is encoded by the FARSB gene.

Phenylalanyl-tRNA synthetase alpha chain is an enzyme that in humans is encoded by the FARSA gene.
Aminoacyl-tRNA synthetases, class II is a family of proteins. These proteins catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have a limited sequence homology.
The aminoacyl-tRNA synthetases catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have limited sequence homology. The 20 aminoacyl-tRNA synthetases are divided into two classes, I and II. Class I aminoacyl-tRNA synthetases contain a characteristic Rossmann fold catalytic domain and are mostly monomeric. Class II aminoacyl-tRNA synthetases share an anti-parallel beta-sheet fold flanked by alpha-helices, and are mostly dimeric or multimeric, containing at least three conserved regions. However, tRNA binding involves an alpha-helical structure that is conserved between class I and class II synthetases. In reactions catalysed by the class I aminoacyl-tRNA synthetases, the aminoacyl group is coupled to the 2'-hydroxyl of the tRNA, while, in class II reactions, the 3'-hydroxyl site is preferred. The synthetases specific for arginine, cysteine, glutamic acid, glutamine, isoleucine, leucine, methionine, tyrosine, tryptophan and valine belong to class I synthetases; these synthetases are further divided into three subclasses, a, b and c, according to sequence homology. The synthetases specific for alanine, asparagine, aspartic acid, glycine, histidine, lysine, phenylalanine, proline, serine, and threonine belong to class-II synthetases.

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.

The FPG IleRS zinc finger domain represents a zinc finger domain found at the C-terminal in both DNA glycosylase/AP lyase enzymes and in isoleucyl tRNA synthetase. In these two types of enzymes, the C-terminal domain forms a zinc finger.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.

In molecular biology, Domain B5 is found in phenylalanine-tRNA synthetase beta subunits. This domain has been shown to bind DNA through a winged helix-turn-helix motif. Phenylalanine-tRNA synthetase may influence common cellular processes via DNA binding, in addition to its aminoacylation function.
















