Related Research Articles
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. Each sequence within an individual prokaryotic cell is derived from a DNA fragment of a bacteriophage that had previously infected the prokaryote or one of its ancestors. These sequences are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes and provide a form of heritable, acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.
Guide RNA (gRNA) or single guide RNA (sgRNA) is a short sequence of RNA that functions as a guide for the Cas9-endonuclease or other Cas-proteins that cut the double-stranded DNA and thereby can be used for gene editing. In bacteria and archaea, gRNAs are a part of the CRISPR-Cas system that serves as an adaptive immune defense that protects the organism from viruses. Here the short gRNAs serve as detectors of foreign DNA and direct the Cas-enzymes that degrades the foreign nucleic acid.

The restriction endonuclease Fok1, naturally found in Flavobacterium okeanokoites, is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non sequence-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves the DNA at two locations, regardless of the nucleotide sequence at the cut site. The DNA is cut 9 nucleotides downstream of the motif on the forward strand, and 13 nucleotides downstream of the motif on the reverse strand, producing two sticky ends with 4-bp overhangs.

In Molecular biology, an insert is a piece of DNA that is inserted into a larger DNA vector by a recombinant DNA technique, such as ligation or recombination. This allows it to be multiplied, selected, further manipulated or expressed in a host organism.

Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain. Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing in situ, a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing.

Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases, and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ).

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.

Feng Zhang is a Chinese–American biochemist. Zhang currently holds the James and Patricia Poitras Professorship in Neuroscience at the McGovern Institute for Brain Research and in the departments of Brain and Cognitive Sciences and Biological Engineering at the Massachusetts Institute of Technology. He also has appointments with the Broad Institute of MIT and Harvard. He is most well known for his central role in the development of optogenetics and CRISPR technologies.

CRISPR interference (CRISPRi) is a genetic perturbation technique that allows for sequence-specific repression of gene expression in prokaryotic and eukaryotic cells. It was first developed by Stanley Qi and colleagues in the laboratories of Wendell Lim, Adam Arkin, Jonathan Weissman, and Jennifer Doudna. Sequence-specific activation of gene expression refers to CRISPR activation (CRISPRa).

Epigenome editing or epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites. Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question.
A protospacer adjacent motif (PAM) is a 2–6-base pair DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system. The PAM is a component of the invading virus or plasmid, but is not found in the bacterial host genome and hence is not a component of the bacterial CRISPR locus. Cas9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence. PAM is an essential targeting component which distinguishes bacterial self from non-self DNA, thereby preventing the CRISPR locus from being targeted and destroyed by the CRISPR-associated nuclease.

Cas12a is an RNA-guided endonuclease that forms an essential component of the CRISPR systems found in some bacteria and archaea. In its natural context, Cas12a targets and destroys the genetic material of viruses and other foreign mobile genetic elements, thereby protecting the host cell from infection. Like other Cas enzymes, Cas12a binds to a "guide" RNA which targets it to a DNA sequence in a specific and programmable matter. In the host organism, the crRNA contains a constant region that is recognized by the Cas12a protein and a "spacer" region that is complementary to a piece of foreign nucleic acid that previously infected the cell.
J. Keith Joung is an American pathologist and molecular biologist who holds the Robert B. Colvin Endowed Chair in Pathology at Massachusetts General Hospital and is Professor of Pathology at Harvard Medical School. He is a leading figure in the field of genome editing and has pioneered the development of designer nucleases and sensitive off-target detection methods.
Off-target genome editing refers to nonspecific and unintended genetic modifications that can arise through the use of engineered nuclease technologies such as: clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9, transcription activator-like effector nucleases (TALEN), meganucleases, and zinc finger nucleases (ZFN). These tools use different mechanisms to bind a predetermined sequence of DNA (“target”), which they cleave, creating a double-stranded chromosomal break (DSB) that summons the cell's DNA repair mechanisms and leads to site-specific modifications. If these complexes do not bind at the target, often a result of homologous sequences and/or mismatch tolerance, they will cleave off-target DSB and cause non-specific genetic modifications. Specifically, off-target effects consist of unintended point mutations, deletions, insertions inversions, and translocations.

CRISPR gene editing (CRISPR, pronounced, refers to a clustered regularly interspaced short palindromic repeats") is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed or new ones added in vivo.
GUIDE-Seq is a molecular biology technique that allows for the unbiased in vitro detection of off-target genome editing events in DNA caused by CRISPR/Cas9 as well as other RNA-guided nucleases in living cells. Similar to LAM-PCR, it employs multiple PCRs to amplify regions of interest that contain a specific insert that preferentially integrates into double-stranded breaks. As gene therapy is an emerging field, GUIDE-Seq has gained traction as a cheap method to detect the off-target effects of potential therapeutics without needing whole genome sequencing.
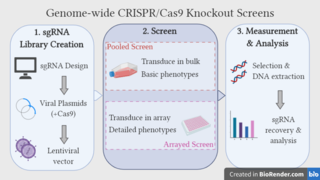
Genome-wide CRISPR-Cas9 knockout screens aim to elucidate the relationship between genotype and phenotype by ablating gene expression on a genome-wide scale and studying the resulting phenotypic alterations. The approach utilises the CRISPR-Cas9 gene editing system, coupled with libraries of single guide RNAs (sgRNAs), which are designed to target every gene in the genome. Over recent years, the genome-wide CRISPR screen has emerged as a powerful tool for performing large-scale loss-of-function screens, with low noise, high knockout efficiency and minimal off-target effects.
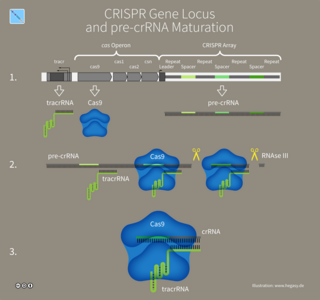
CRISPR RNA or crRNA is a RNA transcript from the CRISPR locus. CRISPR-Cas is an adaptive immune system found in bacteria and archaea to protect against mobile genetic elements, like viruses, plasmids, and transposons. The CRISPR locus contains a series of repeats interspaced with unique spacers. These unique spacers can be acquired from MGEs.
References
- 1 2 3 Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, et al. (December 2014). "Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation". Nature Biotechnology. 32 (12): 1262–7. doi:10.1038/nbt.3026. PMC 4262738 . PMID 25184501.
- 1 2 Chari R, Mali P, Moosburner M, Church GM (September 2015). "Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach". Nature Methods. 12 (9): 823–6. doi:10.1038/nmeth.3473. PMC 5292764 . PMID 26167643.
- ↑ Xiang X, Corsi, GI, Anthon C, Qu K, Pan X, Liang X, Han P, Dong Z, Liu L, Zhong J, Ma T, Wang J, Zhang X, Jiang H, Xu F, Liu X, Xu X, Wang J, Yang H, Bolund L, Church GM, Lin L, Gorodkin J, Luo Y (May 2021). "Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning". Nature Communications. 12 (1): 3238. Bibcode:2021NatCo..12.3238X. doi: 10.1038/s41467-021-23576-0 . PMC 8163799 . PMID 34050182.
- ↑ Alkan F, Wenzel A, Anthon C, Havgaard JH, Gorodkin J (October 2018). "CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters". Genome Biology. 19 (177): 177. doi: 10.1186/s13059-018-1534-x . PMC 6203265 . PMID 30367669.
- ↑ Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD (January 2017). "Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA". Journal of Biotechnology. 241: 136–146. doi: 10.1016/j.jbiotec.2016.11.011 . PMID 27845164.
- ↑ Oliveros JC, Franch M, Tabas-Madrid D, San-León D, Montoliu L, Cubas P, Pazos F (July 2016). "Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes". Nucleic Acids Research. 44 (W1): W267-71. doi:10.1093/nar/gkw407. PMC 4987939 . PMID 27166368.
- ↑ Bae S, Park J, Kim JS (May 2014). "Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases". Bioinformatics. 30 (10): 1473–5. doi:10.1093/bioinformatics/btu048. PMC 4016707 . PMID 24463181.
- ↑ Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F (October 2016). "Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system". Scientific Reports. 6: 35766. Bibcode:2016NatSR...635766E. doi:10.1038/srep35766. PMC 5073330 . PMID 27767081.
- ↑ Pedersen LE, Ronda C, Hansen HG, Kallehauge TB, Betenbaugh MJ, Nielsen AT, Kildegaard HF (August 2014). "Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy". Biotechnology and Bioengineering. 111 (8): 1604–1616. doi:10.1002/bit.25233. PMC 4312910 . PMID 24827782.
- ↑ Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL (2015). "CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool". PLOS ONE. 10 (4): e0124633. Bibcode:2015PLoSO..1024633S. doi: 10.1371/journal.pone.0124633 . PMC 4409221 . PMID 25909470.
- ↑ Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (July 2014). "CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing". Nucleic Acids Research. 42 (Web Server issue): W401-7. doi:10.1093/nar/gku410. PMC 4086086 . PMID 24861617.
- ↑ Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (July 2016). "CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering". Nucleic Acids Research. 44 (W1): W272–6. doi:10.1093/nar/gkw398. PMC 4987937 . PMID 27185894.
- ↑ Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, et al. (July 2016). "Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR". Genome Biology. 17 (1): 148. doi: 10.1186/s13059-016-1012-2 . PMC 4934014 . PMID 27380939.
- ↑ Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. (September 2013). "DNA targeting specificity of RNA-guided Cas9 nucleases". Nature Biotechnology. 31 (9): 827–32. doi:10.1038/nbt.2647. hdl:1721.1/102691. PMC 3969858 . PMID 23873081.
- ↑ Naito Y, Hino K, Bono H, Ui-Tei K (April 2015). "CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites". Bioinformatics. 31 (7): 1120–3. doi:10.1093/bioinformatics/btu743. PMC 4382898 . PMID 25414360.
- ↑ Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ (October 2015). "CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo". Nature Methods. 12 (10): 982–8. doi:10.1038/nmeth.3543. PMC 4589495 . PMID 26322839.
- ↑ Zhu LJ, Holmes BR, Aronin N, Brodsky MH (2014). "CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems". PLOS ONE. 9 (9): e108424. Bibcode:2014PLoSO...9j8424Z. doi: 10.1371/journal.pone.0108424 . PMC 4172692 . PMID 25247697.
- ↑ "Desktop Genetics Announces the Launch of DeskGen Gene Editing Platform". American Laboratory. 2015.
- ↑ Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A (April 2017). "GuideScan software for improved single and paired CRISPR guide RNA design". Nature Biotechnology. 35 (4): 347–349. doi:10.1038/nbt.3804. PMC 5607865 . PMID 28263296.
- ↑ O'Brien A, Bailey TL (September 2014). "GT-Scan: identifying unique genomic targets". Bioinformatics. 30 (18): 2673–5. doi:10.1093/bioinformatics/btu354. PMC 4155256 . PMID 24860161.
- ↑ Pliatsika V, Rigoutsos I (January 2015). ""Off-Spotter": very fast and exhaustive enumeration of genomic lookalikes for designing CRISPR/Cas guide RNAs". Biology Direct. 10 (1): 4. doi: 10.1186/s13062-015-0035-z . PMC 4326336 . PMID 25630343.
- ↑ TechCrunch. "Synthego’s genetic toolkit aims to make CRISPR more accessible | May 2017" Retrieved 23 January 2018.
- ↑ Wilson LO, Reti D, O'Brien AR, Dunne RA, Bauer DC (April 2018). "High Activity Target-Site Identification Using Phenotypic Independent CRISPR-Cas9 Core Functionality". The CRISPR Journal. 1 (2): 182–190. doi:10.1089/crispr.2017.0021. PMID 31021206.
- ↑ Wilson LO, Hetzel S, Pockrandt C, Reinert K, Bauer DC (June 2019). "VARSCOT: variant-aware detection and scoring enables sensitive and personalized off-target detection for CRISPR-Cas9". BMC Biotechnology. 19 (1): 40. doi: 10.1186/s12896-019-0535-5 . PMC 6598273 . PMID 31248401.
- ↑ Poudel R, Rodriguez LT, Reisch CR, Rivers AR (April 2022). "GuideMaker: Software to design CRISPR-Cas guide RNA pools in non-model genomes". GigaScience. 11. doi: 10.1093/gigascience/giac007 . PMC 8975720 . PMID 35365834. S2CID 235700000.
- ↑ Hodgkins A, Farne A, Perera S, Grego T, Parry-Smith DJ, Skarnes WC, Iyer V (September 2015). "WGE: a CRISPR database for genome engineering". Bioinformatics. 31 (18): 3078–3080. doi: 10.1093/bioinformatics/btv308 . PMID 25979474.