
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division.

Cyclin-dependent kinases (CDKs) are a predominant group of serine/threonine protein kinases involved in the regulation of the cell cycle and its progression, ensuring the integrity and functionality of cellular machinery. These regulatory enzymes play a crucial role in the regulation of eukaryotic cell cycle and transcription, as well as DNA repair, metabolism, and epigenetic regulation, in response to several extracellular and intracellular signals. They are present in all known eukaryotes, and their regulatory function in the cell cycle has been evolutionarily conserved. The catalytic activities of CDKs are regulated by interactions with CDK inhibitors (CKIs) and regulatory subunits known as cyclins. Cyclins have no enzymatic activity themselves, but they become active once they bind to CDKs. Without cyclin, CDK is less active than in the cyclin-CDK heterodimer complex. CDKs phosphorylate proteins on serine (S) or threonine (T) residues. The specificity of CDKs for their substrates is defined by the S/T-P-X-K/R sequence, where S/T is the phosphorylation site, P is proline, X is any amino acid, and the sequence ends with lysine (K) or arginine (R). This motif ensures CDKs accurately target and modify proteins, crucial for regulating cell cycle and other functions. Deregulation of the CDK activity is linked to various pathologies, including cancer, neurodegenerative diseases, and stroke.
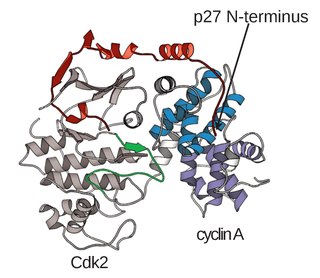
A cyclin-dependent kinase complex is a protein complex formed by the association of an inactive catalytic subunit of a protein kinase, cyclin-dependent kinase (CDK), with a regulatory subunit, cyclin. Once cyclin-dependent kinases bind to cyclin, the formed complex is in an activated state. Substrate specificity of the activated complex is mainly established by the associated cyclin within the complex. Activity of CDKCs is controlled by phosphorylation of target proteins, as well as binding of inhibitory proteins.

S phase (Synthesis phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved.

The restriction point (R), also known as the Start or G1/S checkpoint, is a cell cycle checkpoint in the G1 phase of the animal cell cycle at which the cell becomes "committed" to the cell cycle, and after which extracellular signals are no longer required to stimulate proliferation. The defining biochemical feature of the restriction point is the activation of G1/S- and S-phase cyclin-CDK complexes, which in turn phosphorylate proteins that initiate DNA replication, centrosome duplication, and other early cell cycle events. It is one of three main cell cycle checkpoints, the other two being the G2-M DNA damage checkpoint and the spindle checkpoint.
E2F is a group of genes that encodes a family of transcription factors (TF) in higher eukaryotes. Three of them are activators: E2F1, 2 and E2F3a. Six others act as repressors: E2F3b, E2F4-8. All of them are involved in the cell cycle regulation and synthesis of DNA in mammalian cells. E2Fs as TFs bind to the TTTCCCGC consensus binding site in the target promoter sequence.

Cell cycle checkpoints are control mechanisms in the eukaryotic cell cycle which ensure its proper progression. Each checkpoint serves as a potential termination point along the cell cycle, during which the conditions of the cell are assessed, with progression through the various phases of the cell cycle occurring only when favorable conditions are met. There are many checkpoints in the cell cycle, but the three major ones are: the G1 checkpoint, also known as the Start or restriction checkpoint or Major Checkpoint; the G2/M checkpoint; and the metaphase-to-anaphase transition, also known as the spindle checkpoint. Progression through these checkpoints is largely determined by the activation of cyclin-dependent kinases by regulatory protein subunits called cyclins, different forms of which are produced at each stage of the cell cycle to control the specific events that occur therein.
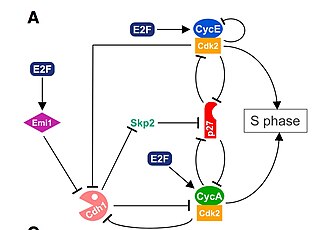
The G1/S transition is a stage in the cell cycle at the boundary between the G1 phase, in which the cell grows, and the S phase, during which DNA is replicated. It is governed by cell cycle checkpoints to ensure cell cycle integrity and the subsequent S phase can pause in response to improperly or partially replicated DNA. During this transition the cell makes decisions to become quiescent, differentiate, make DNA repairs, or proliferate based on environmental cues and molecular signaling inputs. The G1/S transition occurs late in G1 and the absence or improper application of this highly regulated checkpoint can lead to cellular transformation and disease states such as cancer.
The MAPK/ERK pathway is a chain of proteins in the cell that communicates a signal from a receptor on the surface of the cell to the DNA in the nucleus of the cell.
Cyclin A is a member of the cyclin family, a group of proteins that function in regulating progression through the cell cycle. The stages that a cell passes through that culminate in its division and replication are collectively known as the cell cycle Since the successful division and replication of a cell is essential for its survival, the cell cycle is tightly regulated by several components to ensure the efficient and error-free progression through the cell cycle. One such regulatory component is cyclin A which plays a role in the regulation of two different cell cycle stages.
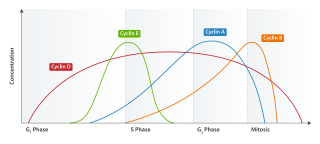
Cyclin E is a member of the cyclin family.

CDK-activating kinase (CAK) activates the cyclin-CDK complex by phosphorylating threonine residue 160 in the CDK activation loop. CAK itself is a member of the Cdk family and functions as a positive regulator of Cdk1, Cdk2, Cdk4, and Cdk6.

Cyclin D is a member of the cyclin protein family that is involved in regulating cell cycle progression. The synthesis of cyclin D is initiated during G1 and drives the G1/S phase transition. Cyclin D protein is anywhere from 155 to 477 amino acids in length.

Cyclin-dependent kinase 2, also known as cell division protein kinase 2, or Cdk2, is an enzyme that in humans is encoded by the CDK2 gene. The protein encoded by this gene is a member of the cyclin-dependent kinase family of Ser/Thr protein kinases. This protein kinase is highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, also known as Cdk1 in humans. It is a catalytic subunit of the cyclin-dependent kinase complex, whose activity is restricted to the G1-S phase of the cell cycle, where cells make proteins necessary for mitosis and replicate their DNA. This protein associates with and is regulated by the regulatory subunits of the complex including cyclin E or A. Cyclin E binds G1 phase Cdk2, which is required for the transition from G1 to S phase while binding with Cyclin A is required to progress through the S phase. Its activity is also regulated by phosphorylation. Multiple alternatively spliced variants and multiple transcription initiation sites of this gene have been reported. The role of this protein in G1-S transition has been recently questioned as cells lacking Cdk2 are reported to have no problem during this transition.

Cyclin-dependent kinase 4 also known as cell division protein kinase 4 is an enzyme that in humans is encoded by the CDK4 gene. CDK4 is a member of the cyclin-dependent kinase family.

Cell division protein kinase 6 (CDK6) is an enzyme encoded by the CDK6 gene. It is regulated by cyclins, more specifically by Cyclin D proteins and Cyclin-dependent kinase inhibitor proteins. The protein encoded by this gene is a member of the cyclin-dependent kinase, (CDK) family, which includes CDK4. CDK family members are highly similar to the gene products of Saccharomyces cerevisiae cdc28, and Schizosaccharomyces pombe cdc2, and are known to be important regulators of cell cycle progression in the point of regulation named R or restriction point.
The Cyclin D/Cdk4 complex is a multi-protein structure consisting of the proteins Cyclin D and cyclin-dependent kinase 4, or Cdk4, a serine-threonine kinase. This complex is one of many cyclin/cyclin-dependent kinase complexes that are the "hearts of the cell-cycle control system" and govern the cell cycle and its progression. As its name would suggest, the cyclin-dependent kinase is only active and able to phosphorylate its substrates when it is bound by the corresponding cyclin. The Cyclin D/Cdk4 complex is integral for the progression of the cell from the Growth 1 phase to the Synthesis phase of the cell cycle, for the Start or G1/S checkpoint.

Cyclin-A2 is a protein that in humans is encoded by the CCNA2 gene. It is one of the two types of cyclin A: cyclin A1 is expressed during meiosis and embryogenesis while cyclin A2 is expressed in the mitotic division of somatic cells.

The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, inactivating it, and the cell cycle is allowed to progress. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases.
The Neuronal cell cycle represents the life cycle of the biological cell, its creation, reproduction and eventual death. The process by which cells divide into two daughter cells is called mitosis. Once these cells are formed they enter G1, the phase in which many of the proteins needed to replicate DNA are made. After G1, the cells enter S phase during which the DNA is replicated. After S, the cell will enter G2 where the proteins required for mitosis to occur are synthesized. Unlike most cell types however, neurons are generally considered incapable of proliferating once they are differentiated, as they are in the adult nervous system. Nevertheless, it remains plausible that neurons may re-enter the cell cycle under certain circumstances. Sympathetic and cortical neurons, for example, try to reactivate the cell cycle when subjected to acute insults such as DNA damage, oxidative stress, and excitotoxicity. This process is referred to as “abortive cell cycle re-entry” because the cells usually die in the G1/S checkpoint before DNA has been replicated.

















