
Cysteine is a semiessential proteinogenic amino acid with the formula HOOC−CH(−NH2)−CH2−SH. The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile. Cysteine is chiral, only L-cysteine is found in nature.

Methionine is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. It is encoded by the codon AUG.

Post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes translating mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).

Cystathionine-β-synthase, also known as CBS, is an enzyme (EC 4.2.1.22) that in humans is encoded by the CBS gene. It catalyzes the first step of the transsulfuration pathway, from homocysteine to cystathionine:
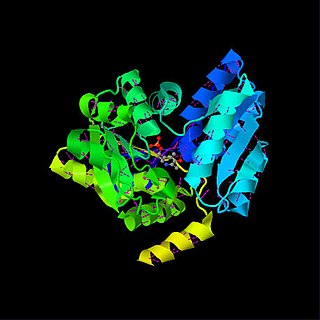
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structural and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes is the deamination of L-serine to yield pyruvate, with the release of ammonia.
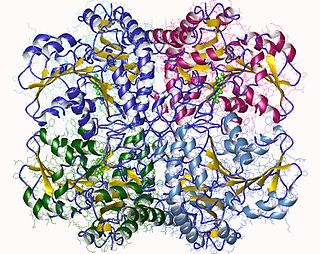
The enzyme cystathionine γ-lyase (EC 4.4.1.1, CTH or CSE; also cystathionase; systematic name L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming)) breaks down cystathionine into cysteine, 2-oxobutanoate (α-ketobutyrate), and ammonia:

The transsulfuration pathway is a metabolic pathway involving the interconversion of cysteine and homocysteine through the intermediate cystathionine. Two transsulfurylation pathways are known: the forward and the reverse.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

The enzyme methionine γ-lyase (EC 4.4.1.11, MGL) is in the γ-family of PLP-dependent enzymes. It degrades sulfur-containing amino acids to α-keto acids, ammonia, and thiols:

Threonine ammonia-lyase (EC 4.3.1.19, systematic name L-threonine ammonia-lyase (2-oxobutanoate-forming), also commonly referred to as threonine deaminase or threonine dehydratase, is an enzyme responsible for catalyzing the conversion of L-threonine into α-ketobutyrate and ammonia:

In enzymology, a serine C-palmitoyltransferase (EC 2.3.1.50) is an enzyme that catalyzes the chemical reaction:

In enzymology, a serine O-acetyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a cystathionine gamma-synthase is an enzyme that catalyzes the formation of cystathionine from cysteine and an activated derivative of homoserine, e.g.:
In enzymology, a cysteine synthase is an enzyme that catalyzes the chemical reaction
In enzymology, an O-acetylhomoserine aminocarboxypropyltransferase is an enzyme that catalyzes the chemical reaction

In biochemistry, non-coded or non-proteinogenic amino acids are distinct from the 22 proteinogenic amino acids which are naturally encoded in the genome of organisms for the assembly of proteins. However, over 140 non-proteinogenic amino acids occur naturally in proteins and thousands more may occur in nature or be synthesized in the laboratory. Chemically synthesized amino acids can be called unnatural amino acids. Unnatural amino acids can be synthetically prepared from their native analogs via modifications such as amine alkylation, side chain substitution, structural bond extension cyclization, and isosteric replacements within the amino acid backbone. Many non-proteinogenic amino acids are important:
Glutamate 2,3-aminomutase is an enzyme that belongs to the radical s-adenosyl methionine (SAM) superfamily. Radical SAM enzymes facilitate the reductive cleavage of S-adenosylmethionine (SAM) through the use of radical chemistry and an iron-sulfur cluster. This enzyme family is implicated in the biosynthesis of DNA precursors, vitamin, cofactor, antibiotic and herbicides and in biodegradation pathways. In particular, glutamate 2,3 aminomutase is involved in the conversion of L-alpha-glutamate to L-beta-glutamate in Clostridium difficile. The generalized reaction is shown below:
















