
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA.
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.
Deoxyribonuclease refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families, which differ in their substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include purifying proteins when extracted from prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Assays of DNase are emerging in the research field as well.

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
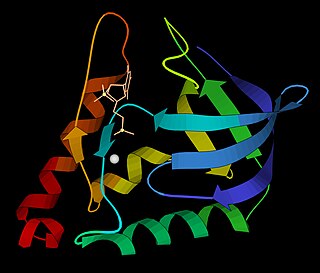
Micrococcal nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved.

Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of the AP site created when DNA glycosylase removes the damaged base.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
Mung bean nuclease is a nuclease derived from sprouts of the mung bean that removes nucleotides in a step-wise manner from single-stranded DNA molecules (ssDNA) and is used in biotechnological applications to remove such ssDNA from a mixture also containing double-stranded DNA (dsDNA). This enzyme is useful for transcript mapping, removal of single-stranded regions in DNA hybrids or single-stranded overhangs produced by restriction enzymes, etc. It has an activity similar to Nuclease S1, but it has higher specificity for single-stranded molecules.
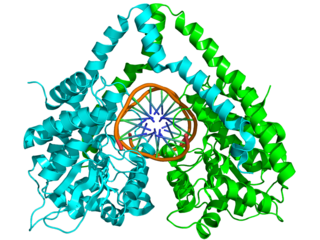
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.
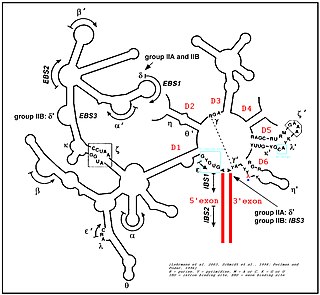
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and deliver cargo DNA such as reporter genes or lox sites

Nuclease S1 is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

BglII is a type II restriction endonuclease isolated from certain strains of Bacillus globigii.

Branch migration is the process by which base pairs on homologous DNA strands are consecutively exchanged at a Holliday junction, moving the branch point up or down the DNA sequence. Branch migration is the second step of genetic recombination, following the exchange of two single strands of DNA between two homologous chromosomes. The process is random, and the branch point can be displaced in either direction on the strand, influencing the degree of which the genetic material is exchanged. Branch migration can also be seen in DNA repair and replication, when filling in gaps in the sequence. It can also be seen when a foreign piece of DNA invades the strand.
Serratia marcescens nuclease is an enzyme. This enzyme catalyses the following chemical reaction

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.

The first step of transcription for some negative, single-stranded RNA viruses is cap snatching, in which the first 10 to 20 residues of a host cell RNA are removed (snatched) and used as the 5′ cap and primer to initiate the synthesis of the nascent viral mRNA. The viral RNA-dependent RNA polymerase (RdRp) can then proceed to replicate the negative-sense genome from the positive-sense template. Cap-snatching also explains why some viral mRNA have 5’ terminal extensions of 10-20 nucleotides that are not encoded for in the genome. Examples of viruses that engage in cap-snatching include influenza viruses (Orthomyxoviridae), Lassa virus (Arenaviridae), hantaan virus (Hantaviridae) and rift valley fever virus (Phenuiviridae). Most viruses snatch 15-20 nucleotides except for the families Arenaviridae and Nairoviridae and the genus Thogotovirus (Orthomyxoviridae) which use a shorter strand.












