In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 um) of 30 nm diameter chromatin fibers.

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.

Saccharomyces cerevisiae is a species of yeast. The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. It is the microorganism behind the most common type of fermentation. S. cerevisiae cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding.

Autophagy is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly.

A fungal prion is a prion that infects fungal hosts. Fungal prions are naturally occurring proteins that can switch between multiple, structurally distinct conformations, at least one of which is self-propagating and transmissible to other prions. This transmission of protein state represents an epigenetic phenomenon where information is encoded in the protein structure itself, instead of in nucleic acids. Several prion-forming proteins have been identified in fungi, primarily in the yeast Saccharomyces cerevisiae. These fungal prions are generally considered benign, and in some cases even confer a selectable advantage to the organism.
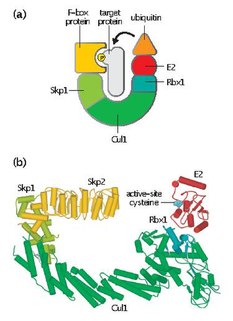
Skp, Cullin, F-box containing complex is a multi-protein E3 ubiquitin ligase complex that catalyzes the ubiquitination of proteins destined for 26S proteasomal degradation. Along with the anaphase-promoting complex, SCF has important roles in the ubiquitination of proteins involved in the cell cycle. The SCF complex also marks various other cellular proteins for destruction.
The Ty5 is a type of retrotransposon native to the Saccharomyces cerevisiae organism.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.

TRAMP complex is a multiprotein, heterotrimeric complex having distributive polyadenylation activity and identifies wide varieties of RNAs produced by polymerases. It was originally discovered in Saccharomycescerevisiae by LaCava et al., Vanacova et al. and Wyers et al. in 2005.
TEAD2, together with TEAD1, defines a novel family of transcription factors, the TEAD family, highly conserved through evolution. TEAD proteins were notably found in Drosophila (Scalloped), C. elegans, S. cerevisiae and A. nidulans. TEAD2 has been less studied than TEAD1 but a few studies revealed its role during development.
Regulator of nonsense transcripts 1 is a protein that in humans is encoded by the UPF1 gene.

Homeobox protein DLX-5 is a protein that in humans is encoded by the distal-less homeobox 5 gene, or DLX5 gene. DLX5 is a member of DLX gene family.

F-box/WD repeat-containing protein 4 is a protein that in humans is encoded by the FBXW4 gene.
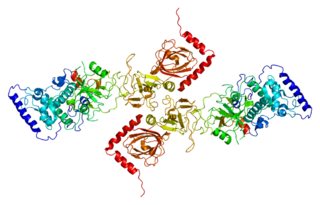
26S proteasome complex subunit DSS1 is a protein that in humans is encoded by the SHFM1 gene.
A killer yeast is a yeast, such as Saccharomyces cerevisiae, which is able to secrete one of a number of toxic proteins which are lethal to susceptible cells. These "killer toxins" are polypeptides that kill sensitive cells of the same or related species, often functioning by creating pores in target cell membranes. These yeast cells are immune to the toxic effects of the protein due to an intrinsic immunity. Killer yeast strains can be a problem in commercial processing because they can kill desirable strains. The killer yeast system was first described in 1963. Study of killer toxins helped to better understand the secretion pathway of yeast, which is similar to those of more complex eukaryotes. It also can be used in treatment of some diseases, mainly those caused by fungi.

Wee1 is a nuclear kinase belonging to the Ser/Thr family of protein kinases in the fission yeast Schizosaccharomyces pombe. Wee1 has a molecular mass of 96 kDa and is a key regulator of cell cycle progression. It influences cell size by inhibiting the entry into mitosis, through inhibiting Cdk1. Wee1 has homologues in many other organisms, including mammals.

Cdc4 is a substrate recognition component of the SCF ubiquitin ligase complex, which acts as a mediator of ubiquitin transfer to target proteins, leading to their subsequent degradation via the ubiquitin-proteasome pathway. Cdc4 targets primarily cell cycle regulators for proteolysis. It serves the function of an adaptor that brings target molecules to the core SCF complex. Cdc4 was originally identified in the model organism Saccharomyces cerevisiae. CDC4 gene function is required at G1/S and G2/M transitions during mitosis and at various stages during meiosis.
Cryptic unstable transcripts (CUTs) are a subset of non-coding RNAs (ncRNAs) that are produced from intergenic and intragenic regions. CUTs were first observed in S. cerevisiae yeast models and are found in most eukaryotes. Some basic characteristics of CUTs include a length of around 200–800 base pairs, a 5' cap, poly-adenylated tail, and rapid degradation due to the combined activity of poly-adenylating polymerases and exosome complexes. CUT transcription occurs through RNA Polymerase II and initiates from nucleosome-depleted regions, often in an antisense orientation. To date, CUTs have a relatively uncharacterized function but have been implicated in a number of putative gene regulation and silencing pathways. Thousands of loci leading to the generation of CUTs have been described in the yeast genome. Additionally, stable uncharacterized transcripts, or SUTs, have also been detected in cells and bear many similarities to CUTs but are not degraded through the same pathways.
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast Saccharomyces cerevisiae e.g. Oaf1, Pip2, Pdr1, Pdr3, Leu3.

Joseph Heitman is an American physician-scientist focused on research in genetics, microbiology, and infectious diseases. He is the James B. Duke Professor and Chair of the Department of Molecular Genetics and Microbiology at Duke University School of Medicine.