
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

In genetics, a deletion is a mutation in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency.

The Philadelphia chromosome or Philadelphia translocation (Ph) is a specific genetic abnormality in chromosome 22 of leukemia cancer cells. This chromosome is defective and unusually short because of reciprocal translocation, t(9;22)(q34;q11), of genetic material between chromosome 9 and chromosome 22, and contains a fusion gene called BCR-ABL1. This gene is the ABL1 gene of chromosome 9 juxtaposed onto the breakpoint cluster region BCR gene of chromosome 22, coding for a hybrid protein: a tyrosine kinase signaling protein that is "always on", causing the cell to divide uncontrollably by interrupting the stability of the genome and impairing various signaling pathways governing the cell cycle.

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal-, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously.

Miller–Dieker syndrome, Miller–Dieker lissencephaly syndrome (MDLS), and chromosome 17p13.3 deletion syndrome is a micro deletion syndrome characterized by congenital malformations. Congenital malformations are physical defects detectable in an infant at birth which can involve many different parts of the body including the brain, hearts, lungs, liver, bones, or intestinal tract. MDS is a contiguous gene syndrome – a disorder due to the deletion of multiple gene loci adjacent to one another. The disorder arises from the deletion of part of the small arm of chromosome 17p, leading to partial monosomy. There may be unbalanced translocations, or the presence of a ring chromosome 17.
Comparative genomic hybridization(CGH) is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions. This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resolution of 5–10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.

Robertsonian translocation (ROB) is a chromosomal abnormality wherein a certain type of a chromosome becomes attached to another. It is the most common form of chromosomal translocation in humans, affecting 1 out of every 1,000 babies born. It does not usually cause health difficulties, but can in some cases result in genetic disorders such as Down syndrome and Patau syndrome. Robertsonian translocations result in a reduction in the number of chromosomes.

Cri du chat syndrome is a rare genetic disorder due to a partial chromosome deletion on chromosome 5. Its name is a French term referring to the characteristic cat-like cry of affected children. It was first described by Jérôme Lejeune in 1963. The condition affects an estimated 1 in 50,000 live births across all ethnicities and is more common in females by a 4:3 ratio.

Jacobsen syndrome is a rare chromosomal disorder resulting from deletion of genes from chromosome 11 that includes band 11q24.1. It is a congenital disorder. Since the deletion takes place on the q arm of chromosome 11, it is also called 11q terminal deletion disorder. The deletion may range from 5 million to 16 million deleted DNA base pairs. The severity of symptoms depends on the number of deletions; the more deletions there are, the more severe the symptoms are likely to be.

A small supernumerary marker chromosome (sSMC) is an abnormal extra chromosome. It contains copies of parts of one or more normal chromosomes and like normal chromosomes is located in the cell's nucleus, is replicated and distributed into each daughter cell during cell division, and typically has genes which may be expressed. However, it may also be active in causing birth defects and neoplasms. The sSMC's small size makes it virtually undetectable using classical cytogenetic methods: the far larger DNA and gene content of the cell's normal chromosomes obscures those of the sSMC. Newer molecular techniques such as fluorescence in situ hybridization, next generation sequencing, comparative genomic hybridization, and highly specialized cytogenetic G banding analyses are required to study it. Using these methods, the DNA sequences and genes in sSMCs are identified and help define as well as explain any effect(s) it may have on individuals.
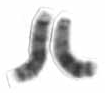
Chromosome 13 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 13 spans about 114 million base pairs and represents between 3.5 and 4% of the total DNA in cells.

Chromosome 22 is one of the 23 pairs of chromosomes in human cells. Humans normally have two copies of chromosome 22 in each cell. Chromosome 22 is the second smallest human chromosome, spanning about 51 million DNA base pairs and representing between 1.5 and 2% of the total DNA in cells.

Chromosome 15 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 15 spans about 99.7 million base pairs and represents between 3% and 3.5% of the total DNA in cells. Chromosome 15 is an acrocentric chromosome, with a very small short arm, which contains few protein coding genes among its 19 million base pairs. It has a larger long arm that is gene rich, spanning about 83 million base pairs.

22q13 deletion syndrome, also known as Phelan–McDermid syndrome (PMS), is a genetic disorder caused by deletions or rearrangements on the q terminal end of chromosome 22. Any abnormal genetic variation in the q13 region that presents with significant manifestations (phenotype) typical of a terminal deletion may be diagnosed as 22q13 deletion syndrome. There is disagreement among researchers as to the exact definition of 22q13 deletion syndrome. The Developmental Synaptopathies Consortium defines PMS as being caused by SHANK3 mutations, a definition that appears to exclude terminal deletions. The requirement to include SHANK3 in the definition is supported by many but not by those who first described 22q13 deletion syndrome.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder, is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.

Acute megakaryoblastic leukemia (AMKL) is life-threatening leukemia in which malignant megakaryoblasts proliferate abnormally and injure various tissues. Megakaryoblasts are the most immature precursor cells in a platelet-forming lineage; they mature to promegakaryocytes and, ultimately, megakaryocytes which cells shed membrane-enclosed particles, i.e. platelets, into the circulation. Platelets are critical for the normal clotting of blood. While malignant megakaryoblasts usually are the predominant proliferating and tissue-damaging cells, their similarly malignant descendants, promegakaryocytes and megakaryocytes, are variable contributors to the malignancy.
Virtual karyotype is the digital information reflecting a karyotype, resulting from the analysis of short sequences of DNA from specific loci all over the genome, which are isolated and enumerated. It detects genomic copy number variations at a higher resolution for level than conventional karyotyping or chromosome-based comparative genomic hybridization (CGH). The main methods used for creating virtual karyotypes are array-comparative genomic hybridization and SNP arrays.
1q21.1 deletion syndrome is a rare aberration of chromosome 1. A human cell has one pair of identical chromosomes on chromosome 1. With the 1q21.1 deletion syndrome, one chromosome of the pair is not complete, because a part of the sequence of the chromosome is missing. One chromosome has the normal length and the other is too short.

1q21.1 duplication syndrome or 1q21.1 (recurrent) microduplication is a rare aberration of chromosome 1.

Chromothripsis is a mutational process by which up to thousands of clustered chromosomal rearrangements occur in a single event in localised and confined genomic regions in one or a few chromosomes, and is known to be involved in both cancer and congenital diseases. It occurs through one massive genomic rearrangement during a single catastrophic event in the cell's history. It is believed that for the cell to be able to withstand such a destructive event, the occurrence of such an event must be the upper limit of what a cell can tolerate and survive. The chromothripsis phenomenon opposes the conventional theory that cancer is the gradual acquisition of genomic rearrangements and somatic mutations over time.
An International System for Human Cytogenetic Nomenclature, Shaffer, L.G., Tommerup N. (eds); S. Karger, Basel 2005














