An Error has occurred retrieving Wikidata item for infobox Fibroblast growth factor 2 (FGF-2), also known as basic fibroblast growth factor (bFGF) and FGF-β, is a growth factor and signaling protein encoded by the FGF2 gene. It binds to and exerts effects via specific fibroblast growth factor receptor (FGFR) proteins, themselves a family of closely related molecules. Fibroblast growth factor protein was first purified in 1975; soon thereafter three variants were isolated: 'basic FGF' (FGF2); Heparin-binding growth factor-2; and Endothelial cell growth factor-2. Gene sequencing revealed that this group is the same FGF2 protein and is a member of a family of FGF proteins.
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. Initiation of eukaryotic translation nearly always occurs at and is dependent on the 5' cap of mRNA molecules, where the translation initiation complex forms and ribosomes engage the mRNA. IRES elements, however allow ribosomes to engage the mRNA and begin translation independently of the 5' cap.
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
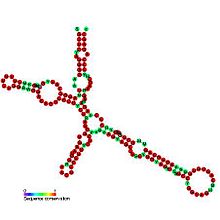
The FGF-2 internal ribosome entry site is an RNA element present in the 5' UTR of the mRNA of fibroblast growth factor-2. It has been found that the FGF-2 internal ribosome entry site (IRES) activity is strictly controlled and highly tissue specific. It is thought that translational IRES dependent activation of FGF-2 plays a vital role in embryogenesis and in the adult brain [1]. When expressed the fibroblast growth factor 2 FGF-2 protein plays a pivotal role in cell proliferation, differentiation and survival as well as being involved in wound-healing [1,2].

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.
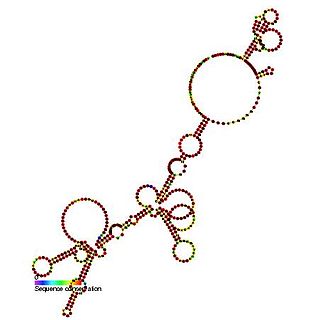
The Hepatitis C virus internal ribosome entry site, or HCV IRES, is an RNA structure within the 5'UTR of the HCV genome that mediates cap-independent translation initiation.
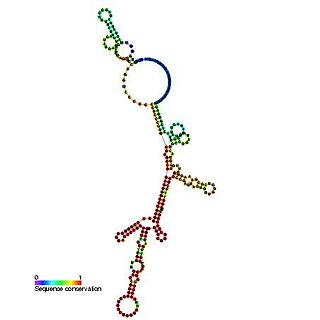
The insulin-like growth factor II (IGF-II) internal ribosome entry site IRES is found in the 5' UTR of IGF-II leader 2 mRNA. This RNA element allows cap-independent translation of the mRNA and it is thought that this family may facilitate a continuous IGF-II production in rapidly dividing cells during development. Ribosomal scanning on human insulin-like growth factor II (IGF-II) is hard to comprehend due to one open reading frame and the ability for the hormone to fold into a stable structure.

The L-myc internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of L-myc that allows cap-independent translation. L-myc undergoes translation via the internal ribosome entry site and bypasses the typical eukaryotic cap-dependent translation pathway [1]. The myc family of genes when expressed are known to be involved in the control of cell growth, differentiation and apoptosis.

The N-myc internal ribosome entry site (IRES) is an RNA element found in the n-myc gene. The myc family of genes when expressed are known to be involved in the control of cell growth, differentiation and apoptosis. n-myc mRNA has an alternative method of translation via an internal ribosome entry site where ribosomes are recruited to the IRES located in the 5' UTR thus bypassing the typical eukaryotic cap-dependent translation pathway.

The HIF-1α internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of HIF-1α that allows cap-independent translation. The HIF-1α internal ribosome entry site (IRES) allows translation to be maintained under hypoxic cell conditions that inhibit cap-dependent translation [1]. The hypoxia-inducible factor-1α protein (HIF-1α) is a subunit of the HIF-1 transcription factor, which induces transcription of several genes involved in the cellular response to hypoxia.
The Tobamovirus internal ribosome entry site (IRES) is an element that allows cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 43S pre-initiation complex directly to the initiation codon and eliminates the requirement for the eukaryotic initiation factor, eIF4F.

This family represents the vascular endothelial growth factor (VEGF) internal ribosome entry site (IRES) A. VEGF is an endothelial cell mitogen with many crucial functions such as embryogenic development and wound healing. The 5' UTR of VEGF mRNA contains two IRES elements which are able to promote efficient translation at the AUG start codon, this family represents IRES A.
A ribosome binding site, or ribosomal binding site (RBS), is a sequence of nucleotides upstream of the start codon of an mRNA transcript that is responsible for the recruitment of a ribosome during the initiation of translation. Mostly, RBS refers to bacterial sequences, although internal ribosome entry sites (IRES) have been described in mRNAs of eukaryotic cells or viruses that infect eukaryotes. Ribosome recruitment in eukaryotes is generally mediated by the 5' cap present on eukaryotic mRNAs.

Keratinocyte growth factor is a protein that in humans is encoded by the FGF7 gene.

Fibroblast growth factor 10 is a protein that in humans is encoded by the FGF10 gene.

Fibroblast growth factor 4 is a protein that in humans is encoded by the FGF4 gene.

Fibroblast growth factor 18 (FGF-18) is a protein that is encoded by the FGF18 gene in humans. The protein was first discovered in 1998, when two newly-identified murine genes Fgf17 and Fgf18 were described and confirmed as being closely related by sequence homology to Fgf8. The three proteins were eventually grouped into the FGF8 subfamily, which contains several of the endocrine FGF superfamily members FGF8, FGF17, and FGF18. Subsequent studies identified FGF18's role in promoting chondrogenesis, and an apparent specific activity for the generation of the hyaline cartilage in articular joints.

Fibroblast growth factor 17 is a protein that in humans is encoded by the FGF17 gene.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human EIF4G1 has been the focus of extensive studies.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.