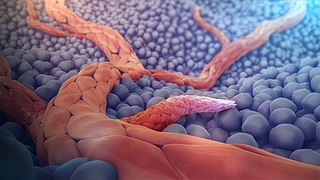
Angiogenesis is the physiological process through which new blood vessels form from pre-existing vessels, formed in the earlier stage of vasculogenesis. Angiogenesis continues the growth of the vasculature by processes of sprouting and splitting. Vasculogenesis is the embryonic formation of endothelial cells from mesoderm cell precursors, and from neovascularization, although discussions are not always precise. The first vessels in the developing embryo form through vasculogenesis, after which angiogenesis is responsible for most, if not all, blood vessel growth during development and in disease.

FGF2, also known as basic fibroblast growth factor (bFGF) and FGF-β, is a growth factor and signaling protein encoded by the FGF2 gene. It is synthesized primarily as a 155 amino acid polypeptide, resulting in an 18 kDa protein. However, there are four alternate start codons which provide N-terminal extensions of 41, 46, 55, or 133 amino acids, resulting in proteins of 22 kDa, 22.5 kDa, 24 kDa and 34 kDa, respectively. Generally, the 155 aa/18 kDa low molecular weight (LMW) form is considered cytoplasmic and can be secreted from the cell, whereas the high molecular weight (HMW) forms are directed to the cell's nucleus.

FGF1, also known as acidic fibroblast growth factor (aFGF), is a growth factor and signaling protein encoded by the FGF1 gene. It is synthesized as a 155 amino acid polypeptide, whose mature form is a non-glycosylated 17-18 kDa protein. Fibroblast growth factor protein was first purified in 1975, but soon afterwards others using different conditions isolated acidic FGF, Heparin-binding growth factor-1, and Endothelial cell growth factor-1. Gene sequencing revealed that this group was actually the same growth factor and that FGF1 was a member of a family of FGF proteins.

The FGF-1 internal ribosome entry site (IRES) is an RNA element present in the 5' UTR of the mRNA of fibroblast growth factor-1 and allows cap-independent translation. It is thought that FGF-1 internal ribosome entry site (IRES) activity is strictly controlled and highly tissue specific.
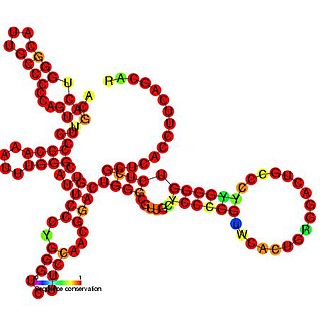
The insulin-like growth factor II (IGF-II) internal ribosome entry site IRES is found in the 5' UTR of IGF-II leader 2 mRNA. This RNA element allows cap-independent translation of the mRNA and it is thought that this family may facilitate a continuous IGF-II production in rapidly dividing cells during development. Ribosomal scanning on human insulin-like growth factor II (IGF-II) is hard to comprehend due to one open reading frame and the ability for the hormone to fold into a stable structure.

This family represents the vascular endothelial growth factor (VEGF) internal ribosome entry site (IRES) A. VEGF is an endothelial cell mitogen with many crucial functions such as embryogenic development and wound healing. The 5' UTR of VEGF mRNA contains two IRES elements which are able to promote efficient translation at the AUG start codon, this family represents IRES A.

Keratinocyte growth factor is a protein that in humans is encoded by the FGF7 gene.

Fibroblast growth factor 10 is a protein that in humans is encoded by the FGF10 gene.

Fibroblast growth factor 8 is a protein that in humans is encoded by the FGF8 gene.

Glia-activating factor is a protein that in humans is encoded by the FGF9 gene.

Fibroblast growth factor 4 is a protein that in humans is encoded by the FGF4 gene.

Fibroblast growth factor 5 is a protein that in humans is encoded by the FGF5 gene.

Fibroblast growth factor 18 is a protein that in humans is encoded by the FGF18 gene.

Fibroblast growth factor 13 is a protein that in humans is encoded by the FGF13 gene.

Fibroblast growth factor 6 is a protein that in humans is encoded by the FGF6 gene.

Fibroblast growth factor 21 is a protein that in mammals is encoded by the FGF21 gene. The protein encoded by this gene is a member of the fibroblast growth factor (FGF) family and specifically a member of the endocrine subfamily which includes FGF23 and FGF15/19. FGF21 is the primary endogenous agonist of the FGF21 receptor, which is composed of the co-receptors FGF receptor 1 and β-Klotho.
Eukaryotic translation initiation factor 4 G (eIF4G) is a protein involved in eukaryotic translation initiation and is a component of the eIF4F cap-binding complex. Orthologs of eIF4G have been studied in multiple species, including humans, yeast, and wheat. However, eIF4G is exclusively found in domain Eukarya, and not in domains Bacteria or Archaea, which do not have capped mRNA. As such, eIF4G structure and function may vary between species, although the human eIF4G 1 has been the focus of extensive studies.
Fibroblast growth factor 22 is a protein which in humans is encoded by the FGF22 gene.

















