The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division.

The G1 phase, gap 1 phase, or growth 1 phase, is the first of four phases of the cell cycle that takes place in eukaryotic cell division. In this part of interphase, the cell synthesizes mRNA and proteins in preparation for subsequent steps leading to mitosis. G1 phase ends when the cell moves into the S phase of interphase. Around 30 to 40 percent of cell cycle time is spent in the G1 phase.

S phase (Synthesis phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S-phase are tightly regulated and widely conserved.

G2 phase, Gap 2 phase, or Growth 2 phase, is the third subphase of interphase in the cell cycle directly preceding mitosis. It follows the successful completion of S phase, during which the cell’s DNA is replicated. G2 phase ends with the onset of prophase, the first phase of mitosis in which the cell’s chromatin condenses into chromosomes.

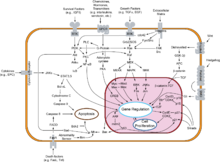
The restriction point (R), also known as the Start or G1/S checkpoint, is a cell cycle checkpoint in the G1 phase of the animal cell cycle at which the cell becomes "committed" to the cell cycle, and after which extracellular signals are no longer required to stimulate proliferation. The defining biochemical feature of the restriction point is the activation of G1/S- and S-phase cyclin-CDK complexes, which in turn phosphorylate proteins that initiate DNA replication, centrosome duplication, and other early cell cycle events. It is one of three main cell cycle checkpoints, the other two being the G2-M DNA damage checkpoint and the spindle checkpoint.
E2F is a group of genes that encodes a family of transcription factors (TF) in higher eukaryotes. Three of them are activators: E2F1, 2 and E2F3a. Six others act as repressors: E2F3b, E2F4-8. All of them are involved in the cell cycle regulation and synthesis of DNA in mammalian cells. E2Fs as TFs bind to the TTTCCCGC consensus binding site in the target promoter sequence.

Cell cycle checkpoints are control mechanisms in the eukaryotic cell cycle which ensure its proper progression. Each checkpoint serves as a potential termination point along the cell cycle, during which the conditions of the cell are assessed, with progression through the various phases of the cell cycle occurring only when favorable conditions are met. There are many checkpoints in the cell cycle, but the three major ones are: the G1 checkpoint, also known as the Start or restriction checkpoint or Major Checkpoint; the G2/M checkpoint; and the metaphase-to-anaphase transition, also known as the spindle checkpoint. Progression through these checkpoints is largely determined by the activation of cyclin-dependent kinases by regulatory protein subunits called cyclins, different forms of which are produced at each stage of the cell cycle to control the specific events that occur therein.
Cyclin A is a member of the cyclin family, a group of proteins that function in regulating progression through the cell cycle. The stages that a cell passes through that culminate in its division and replication are collectively known as the cell cycle Since the successful division and replication of a cell is essential for its survival, the cell cycle is tightly regulated by several components to ensure the efficient and error-free progression through the cell cycle. One such regulatory component is cyclin A which plays a role in the regulation of two different cell cycle stages.

Cyclin D is a member of the cyclin protein family that is involved in regulating cell cycle progression. The synthesis of cyclin D is initiated during G1 and drives the G1/S phase transition. Cyclin D protein is anywhere from 155 to 477 amino acids in length.

Cyclin-dependent kinase 2, also known as cell division protein kinase 2, or Cdk2, is an enzyme that in humans is encoded by the CDK2 gene. The protein encoded by this gene is a member of the cyclin-dependent kinase family of Ser/Thr protein kinases. This protein kinase is highly similar to the gene products of S. cerevisiae cdc28, and S. pombe cdc2, also known as Cdk1 in humans. It is a catalytic subunit of the cyclin-dependent kinase complex, whose activity is restricted to the G1-S phase of the cell cycle, where cells make proteins necessary for mitosis and replicate their DNA. This protein associates with and is regulated by the regulatory subunits of the complex including cyclin E or A. Cyclin E binds G1 phase Cdk2, which is required for the transition from G1 to S phase while binding with Cyclin A is required to progress through the S phase. Its activity is also regulated by phosphorylation. Multiple alternatively spliced variants and multiple transcription initiation sites of this gene have been reported. The role of this protein in G1-S transition has been recently questioned as cells lacking Cdk2 are reported to have no problem during this transition.

Cyclin-dependent kinase 4 (CDK4), also known as cell division protein kinase 4, is an enzyme that is encoded by the CDK4 gene in humans. CDK4 is a member of the cyclin-dependent kinase family, a group of serine/threonine kinases which regulate the cell cycle. CDK4 regulates the G1/S transition by contributing to the phosphorylation of retinoblastoma (RB) protein, which leads to the release of protein factors like E2F1 that promote S-phase progression. It is regulated by cyclins like cyclin D proteins, regulatory kinases, and cyclin kinase inhibitors (CKIs). Dysregulation of the CDK4 pathway is common in many cancers, and CDK4 is a new therapeutic target in cancer treatment.
The Cyclin D/Cdk4 complex is a multi-protein structure consisting of the proteins Cyclin D and cyclin-dependent kinase 4, or Cdk4, a serine-threonine kinase. This complex is one of many cyclin/cyclin-dependent kinase complexes that are the "hearts of the cell-cycle control system" and govern the cell cycle and its progression. As its name would suggest, the cyclin-dependent kinase is only active and able to phosphorylate its substrates when it is bound by the corresponding cyclin. The Cyclin D/Cdk4 complex is integral for the progression of the cell from the Growth 1 phase to the Synthesis phase of the cell cycle, for the Start or G1/S checkpoint.
A series of biochemical switches control transitions between and within the various phases of the cell cycle. The cell cycle is a series of complex, ordered, sequential events that control how a single cell divides into two cells, and involves several different phases. The phases include the G1 and G2 phases, DNA replication or S phase, and the actual process of cell division, mitosis or M phase. During the M phase, the chromosomes separate and cytokinesis occurs.
The Start point is a major cell cycle checkpoint in yeast, known as the restriction point in multicellular organisms. The Start checkpoint ensures cell-cycle entry even if conditions later become unfavorable. The physiological factors that control passage through the Start checkpoint include external nutrient concentrations, presence of mating factor/ pheromone, forms of stress, and size control.

The retinoblastoma protein is a tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, inactivating it, and the cell cycle is allowed to progress. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases.
Clb5 and Clb6 are B-type, S-phase cyclins in yeast that assist in cell cycle regulation. Clb5 and Clb6 bind and activate Cdk1, and high levels of these cyclins are required for entering S-phase. S-phase cyclin binding to Cdk1 directly stimulates DNA replication as well as progression to the next phase of the cell cycle.

DNA re-replication is an undesirable and possibly fatal occurrence in eukaryotic cells in which the genome is replicated more than once per cell cycle. Rereplication is believed to lead to genomic instability and has been implicated in the pathologies of a variety of human cancers. To prevent rereplication, eukaryotic cells have evolved multiple, overlapping mechanisms to inhibit chromosomal DNA from being partially or fully rereplicated in a given cell cycle. These control mechanisms rely on cyclin-dependent kinase (CDK) activity. DNA replication control mechanisms cooperate to prevent the relicensing of replication origins and to activate cell cycle and DNA damage checkpoints. DNA rereplication must be strictly regulated to ensure that genomic information is faithfully transmitted through successive generations.
Whi5 is a transcriptional regulator in the budding yeast, notably in the G1 phase. It plays an important role in cell size control in G1 phase, similarly with Retinoblastoma (Rb) protein in human, although the two have no similarity in sequence Whi5 is an inhibitor of SBF, which is involved in the transcription of G1-specific genes. Cln3 promotes the disassociation of Whi5 from SBF, and its disassociation results in the transcription of genes needed to enter S phase.
The Neuronal cell cycle represents the life cycle of the biological cell, its creation, reproduction and eventual death. The process by which cells divide into two daughter cells is called mitosis. Once these cells are formed they enter G1, the phase in which many of the proteins needed to replicate DNA are made. After G1, the cells enter S phase during which the DNA is replicated. After S, the cell will enter G2 where the proteins required for mitosis to occur are synthesized. Unlike most cell types however, neurons are generally considered incapable of proliferating once they are differentiated, as they are in the adult nervous system. Nevertheless, it remains plausible that neurons may re-enter the cell cycle under certain circumstances. Sympathetic and cortical neurons, for example, try to reactivate the cell cycle when subjected to acute insults such as DNA damage, oxidative stress, and excitotoxicity. This process is referred to as “abortive cell cycle re-entry” because the cells usually die in the G1/S checkpoint before DNA has been replicated.

The Cyclin E/Cdk2 complex is a structure composed of two proteins, cyclin E and cyclin-dependent kinase 2 (Cdk2). Similar to other cyclin/Cdk complexes, the cyclin E/Cdk2 dimer plays a crucial role in regulating the cell cycle, with this specific complex peaking in activity during the G1/S transition. Once the cyclin and Cdk subunits join together, the complex gets activated, allowing it to phosphorylate and bind to downstream proteins to ultimately promote cell cycle progression. Although cyclin E can bind to other Cdk proteins, its primary binding partner is Cdk2, and the majority of cyclin E activity occurs when it exists as the cyclin E/Cdk2 complex.