See also
- Genetic editing, an approach to scholarly editing of literary texts
Gene editing may refer to:
Gene knockouts are a widely used genetic engineering technique that involves the targeted removal or inactivation of a specific gene within an organism's genome. This can be done through a variety of methods, including homologous recombination, CRISPR-Cas9, and TALENs.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) which stabilizes the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

A germline mutation, or germinal mutation, is any detectable variation within germ cells. Mutations in these cells are the only mutations that can be passed on to offspring, when either a mutated sperm or oocyte come together to form a zygote. After this fertilization event occurs, germ cells divide rapidly to produce all of the cells in the body, causing this mutation to be present in every somatic and germline cell in the offspring; this is also known as a constitutional mutation. Germline mutation is distinct from somatic mutation.

A designer baby is a baby whose genetic makeup has been selected or altered, often to exclude a particular gene or to remove genes associated with disease. This process usually involves analysing a wide range of human embryos to identify genes associated with particular diseases and characteristics, and selecting embryos that have the desired genetic makeup; a process known as preimplantation genetic diagnosis. Screening for single genes is commonly practiced, and polygenic screening is offered by a few companies. Other methods by which a baby's genetic information can be altered involve directly editing the genome before birth, which is not routinely performed and only one instance of this is known to have occurred as of 2019, where Chinese twins Lulu and Nana were edited as embryos, causing widespread criticism.

In genetics, an insertion is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an insertion refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis.
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zinc-finger nucleases to target unique sequences within complex genomes. By taking advantage of endogenous DNA repair machinery, these reagents can be used to precisely alter the genomes of higher organisms. Alongside CRISPR/Cas9 and TALEN, ZFN is a prominent tool in the field of genome editing.

The restriction endonuclease Fok1, naturally found in Flavobacterium okeanokoites, is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non sequence-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves the DNA at two locations, regardless of the nucleotide sequence at the cut site. The DNA is cut 9 nucleotides downstream of the motif on the forward strand, and 13 nucleotides downstream of the motif on the reverse strand, producing two sticky ends with 4-bp overhangs.

Gene targeting is a biotechnological tool used to change the DNA sequence of an organism. It is based on the natural DNA-repair mechanism of Homology Directed Repair (HDR), including Homologous Recombination. Gene targeting can be used to make a range of sizes of DNA edits, from larger DNA edits such as inserting entire new genes into an organism, through to much smaller changes to the existing DNA such as a single base-pair change. Gene targeting relies on the presence of a repair template to introduce the user-defined edits to the DNA. The user will design the repair template to contain the desired edit, flanked by DNA sequence corresponding (homologous) to the region of DNA that the user wants to edit; hence the edit is targeted to a particular genomic region. In this way Gene Targeting is distinct from natural homology-directed repair, during which the ‘natural’ DNA repair template of the sister chromatid is used to repair broken DNA. The alteration of DNA sequence in an organism can be useful in both a research context – for example to understand the biological role of a gene – and in biotechnology, for example to alter the traits of an organism.

In Molecular biology, an insert is a piece of DNA that is inserted into a larger DNA vector by a recombinant DNA technique, such as ligation or recombination. This allows it to be multiplied, selected, further manipulated or expressed in a host organism.
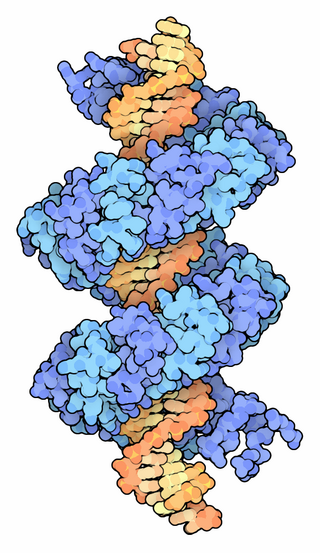
TALeffectors are proteins secreted by some β- and γ-proteobacteria. Most of these are Xanthomonads. Plant pathogenic Xanthomonas bacteria are especially known for TALEs, produced via their type III secretion system. These proteins can bind promoter sequences in the host plant and activate the expression of plant genes that aid bacterial infection. The TALE domain responsible for binding to DNA is known to have 1.5 to 33.5 short sequences that are repeated multiple times. Each of these repeats was found to be specific for a certain base pair of the DNA. These repeats also have repeat variable residues (RVD) that can detect specific DNA base pairs. They recognize plant DNA sequences through a central repeat domain consisting of a variable number of ~34 amino acid repeats. There appears to be a one-to-one correspondence between the identity of two critical amino acids in each repeat and each DNA base in the target sequence. These proteins are interesting to researchers both for their role in disease of important crop species and the relative ease of retargeting them to bind new DNA sequences. Similar proteins can be found in the pathogenic bacterium Ralstonia solanacearum and Burkholderia rhizoxinica, as well as yet unidentified marine microorganisms. The term TALE-likes is used to refer to the putative protein family encompassing the TALEs and these related proteins.
Recombinant adeno-associated virus (rAAV) based genome engineering is a genome editing platform centered on the use of recombinant AAV vectors that enables insertion, deletion or substitution of DNA sequences into the genomes of live mammalian cells. The technique builds on Mario Capecchi and Oliver Smithies' Nobel Prize–winning discovery that homologous recombination (HR), a natural hi-fidelity DNA repair mechanism, can be harnessed to perform precise genome alterations in mice. rAAV mediated genome-editing improves the efficiency of this technique to permit genome engineering in any pre-established and differentiated human cell line, which, in contrast to mouse ES cells, have low rates of HR.

Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain. Transcription activator-like effectors (TALEs) can be engineered to bind to practically any desired DNA sequence, so when combined with a nuclease, DNA can be cut at specific locations. The restriction enzymes can be introduced into cells, for use in gene editing or for genome editing in situ, a technique known as genome editing with engineered nucleases. Alongside zinc finger nucleases and CRISPR/Cas9, TALEN is a prominent tool in the field of genome editing.

Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases, and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ).

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.

Epigenome editing or epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites. Whereas gene editing involves changing the actual DNA sequence itself, epigenetic editing involves modifying and presenting DNA sequences to proteins and other DNA binding factors that influence DNA function. By "editing” epigenomic features in this manner, researchers can determine the exact biological role of an epigenetic modification at the site in question.

Daniel Voytas is an American geneticist who is Professor of Genetics, Cell Biology and Development at the University of Minnesota and Director of the Center for Precision Plant Genomics. He is also the Chief Scientific Officer of Calyxt, an agricultural biotechnology company focused on developing crops that provide consumer benefit.
Off-target genome editing refers to nonspecific and unintended genetic modifications that can arise through the use of engineered nuclease technologies such as: clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9, transcription activator-like effector nucleases (TALEN), meganucleases, and zinc finger nucleases (ZFN). These tools use different mechanisms to bind a predetermined sequence of DNA (“target”), which they cleave, creating a double-stranded chromosomal break (DSB) that summons the cell's DNA repair mechanisms and leads to site-specific modifications. If these complexes do not bind at the target, often a result of homologous sequences and/or mismatch tolerance, they will cleave off-target DSB and cause non-specific genetic modifications. Specifically, off-target effects consist of unintended point mutations, deletions, insertions inversions, and translocations.

CRISPR gene editing standing for "Clustered Regularly Interspaced Short Palindromic Repeats" is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.