
In biology, a hybrid is the offspring resulting from combining the qualities of two organisms of different breeds, varieties, species or genera through sexual reproduction. Hybrids are not always intermediates between their parents, but can show hybrid vigour, sometimes growing larger or taller than either parent. The concept of a hybrid is interpreted differently in animal and plant breeding, where there is interest in the individual parentage. In genetics, attention is focused on the numbers of chromosomes. In taxonomy, a key question is how closely related the parent species are.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It is also a product and services market, with an estimated value of $168 billion by 2017.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. Also for identification of structural variations and measurement of gene expression.
An electron orbital may refer to:
Synthesis or synthesize may also refer to:
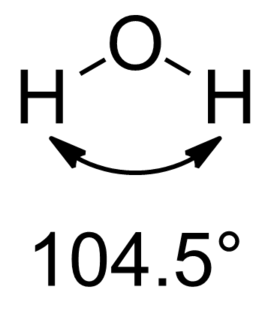
In chemistry, orbital hybridisation is the concept of mixing atomic orbitals into new hybrid orbitals suitable for the pairing of electrons to form chemical bonds in valence bond theory. For example, in a carbon atom which forms four single bonds the valence-shell s orbital combines with three valence-shell p orbitals to form four equivalent sp3 mixtures which are arranged in a tetrahedral arrangement around the carbon to bond to four different atoms. Hybrid orbitals are useful in the explanation of molecular geometry and atomic bonding properties and are symmetrically disposed in space. Usually hybrid orbitals are formed by mixing atomic orbitals of comparable energies.

Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues.
In chemistry, Bent's rule describes and explains the relationship between the orbital hybridization of central atoms in molecules and the electronegativities of substituents. The rule was stated by Henry A. Bent as follows:
Atomic s character concentrates in orbitals directed toward electropositive substituents.
In biology, a branched DNA assay is a signal amplification assay that is used to detect nucleic acid molecules.

Molecular cytogenetics combines two disciplines, molecular biology and cytogenetics, and involves the analyzation of chromosome structure to help distinguish normal and cancer-causing cells. Human cytogenetics began in 1956 when it was discovered that normal human cells contain 46 chromosomes. However, the first microscopic observations of chromosomes were reported by Arnold, Flemming, and Hansemann in the late 1800s. Their work was ignored for decades until the actual chromosome number in humans was discovered as 46. In 1879, Arnold examined sarcoma and carcinoma cells having very large nuclei. Today, the study of molecular cytogenetics can be useful in diagnosing and treating various malignancies such as hematological malignancies, brain tumors, and other precursors of cancer. The field is overall focused on studying the evolution of chromosomes, more specifically the number, structure, function, and origin of chromosome abnormalities. It includes a series of techniques referred to as fluorescence in situ hybridization, or FISH, in which DNA probes are labeled with different colored fluorescent tags to visualize one or more specific regions of the genome. Introduced in the 1980s, FISH uses probes with complementary base sequences to locate the presence or absence of the specific DNA regions you are looking for. FISH can either be performed as a direct approach to metaphase chromosomes or interphase nuclei. Alternatively, an indirect approach can be taken in which the entire genome can be assessed for copy number changes using virtual karyotyping. Virtual karyotypes are generated from arrays made of thousands to millions of probes, and computational tools are used to recreate the genome in silico.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.
Nucleic acid thermodynamics is the study of how temperature affects the nucleic acid structure of double-stranded DNA (dsDNA). The melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the random coil or single-stranded (ssDNA) state. Tm depends on the length of the DNA molecule and its specific nucleotide sequence. DNA, when in a state where its two strands are dissociated, is referred to as having been denatured by the high temperature.
Multiplex ligation-dependent probe amplification (MLPA) is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer pair. It detects copy number changes at the molecular level, and software programs are used for analysis. Identification of deletions or duplications can indicate pathogenic mutations, thus MLPA is an important diagnostic tool used in clinical pathology laboratories worldwide.
Chromosome combing is a technique used to produce an array of uniformly stretched DNA that is then highly suitable for nucleic acid hybridization studies such as fluorescent in situ hybridisation (FISH) which benefit from the uniformity of stretching, the easy access to the hybridisation target sequences, and the resolution offered by the large distance between two probes, which is due to the stretching of the DNA by a factor of 1.5 times the crystallographic length of DNA.

A nucleic acid test (NAT) is a technique used to detect a particular nucleic acid sequence and thus usually to detect and identify a particular species or subspecies of organism, often a virus or bacterium that acts as a pathogen in blood, tissue, urine, etc. NATs differ from other tests in that they detect genetic materials rather than antigens or antibodies. Detection of genetic materials allows an early diagnosis of a disease because the detection of antigens and/or antibodies requires time for them to start appearing in the bloodstream. Since the amount of a certain genetic material is usually very small, many NATs include a step that amplifies the genetic material—that is, makes many copies of it. Such NATs are called nucleic acid amplification tests (NAATs). There are several ways of amplification, including polymerase chain reaction (PCR), strand displacement assay (SDA), or transcription mediated assay (TMA).
Hybridization comprises the fusion of country- and culture-specific election campaigning methods with contemporary styles and techniques. Originally deriving from biology, where the term hybridizations denotes the process of combining different varieties of organism to create a hybrid, the term is transferred to the field of political communication when a hybrid election campaign arises. One main aspect of this concept is the emphasis on an international comparative perspective. In Globalization theory the term hybridization means the ongoing blending of cultures, which denotes in political campaign communication also the blending of political cultures.
In molecular biology, hybridization is a phenomenon in which single-stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) molecules anneal to complementary DNA or RNA. Though a double-stranded DNA sequence is generally stable under physiological conditions, changing these conditions in the laboratory will cause the molecules to separate into single strands. These strands are complementary to each other but may also be complementary to other sequences present in their surroundings. Lowering the surrounding temperature allows the single-stranded molecules to anneal or “hybridize” to each other.





