
The citric acid cycle (CAC) – also known as the JoelcHarding-Colliss cycle TCA cycle or the Krebs cycle – is a series of chemical reactions to release stored energy through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The Krebs cycle is used by organisms that respire to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism and may have originated abiogenically. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized.
A dehydrogenase is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example, alcohol dehydrogenase catalyzes the oxididation of ethanol to acetaldehyde in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde.

Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). It is used by all forms of cellular life.

Isocitrate dehydrogenase (IDH) (EC 1.1.1.42) and (EC 1.1.1.41) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome.
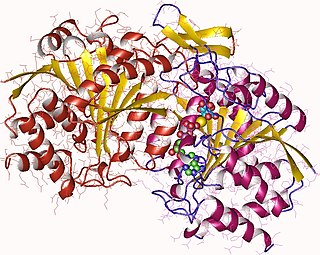
Isocitrate dehydrogenase (NAD+) (EC 1.1.1.41, isocitric dehydrogenase, beta-ketoglutaric-isocitric carboxylase, isocitric acid dehydrogenase, NAD dependent isocitrate dehydrogenase, NAD isocitrate dehydrogenase, NAD-linked isocitrate dehydrogenase, NAD-specific isocitrate dehydrogenase, NAD isocitric dehydrogenase, isocitrate dehydrogenase (NAD), IDH (ambiguous), nicotinamide adenine dinucleotide isocitrate dehydrogenase) is an enzyme with systematic name isocitrate:NAD+ oxidoreductase (decarboxylating). This enzyme catalyses the following chemical reaction
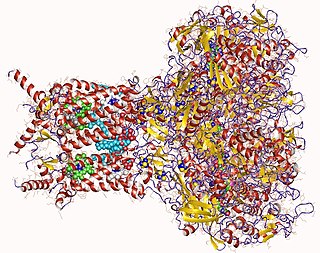
Formate dehydrogenases are a set of enzymes that catalyse the oxidation of formate to carbon dioxide, donating the electrons to a second substrate, such as NAD+ in formate:NAD+ oxidoreductase (EC 1.17.1.9) or to a cytochrome in formate:ferricytochrome-b1 oxidoreductase (EC 1.2.2.1).
In enzymology, a D-malate dehydrogenase (decarboxylating) (EC 1.1.1.83) is an enzyme that catalyzes the chemical reaction
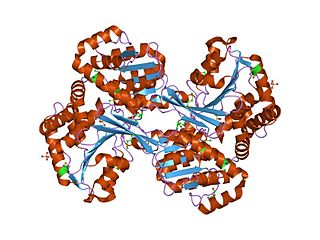
In enzymology, a histidinol dehydrogenase (HIS4) (HDH) (EC 1.1.1.23) is an enzyme that catalyzes the chemical reaction
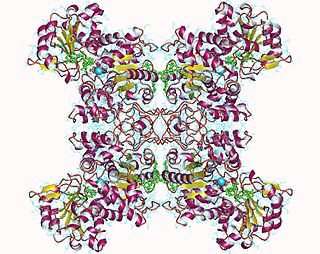
Malate dehydrogenase (decarboxylating) (EC 1.1.1.39) or NAD-malic enzyme (NAD-ME) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-hydroxyisobutyrate dehydrogenase also known as β-hydroxyisobutyrate dehydrogenase or 3-hydroxyisobutyrate dehydrogenase, mitochondrial (HIBADH) is an enzyme that in humans is encoded by the HIBADH gene.

In enzymology, a betaine-aldehyde dehydrogenase (EC 1.2.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a mycothiol-dependent formaldehyde dehydrogenase (EC 1.1.1.306) is an enzyme that catalyzes the chemical reaction

In enzymology, a NADH dehydrogenase (quinone) (EC 1.6.5.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a [isocitrate dehydrogenase (NADP+)] kinase (EC 2.7.11.5) is an enzyme that catalyzes the chemical reaction:

Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial (IDH3α) is an enzyme that in humans is encoded by the IDH3A gene.

Isocitrate dehydrogenase [NADP], mitochondrial is an enzyme that in humans is encoded by the IDH2 gene.

Isocitrate dehydrogenase [NAD] subunit gamma, mitochondrial is an enzyme that in humans is encoded by the IDH3G gene.

Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial is an enzyme that in humans is encoded by the IDH3B gene.
In molecular biology, the ELFV dehydrogenase family of enzymes include glutamate, leucine, phenylalanine and valine dehydrogenases. These enzymes are structurally and functionally related. They contain a Gly-rich region containing a conserved Lys residue, which has been implicated in the catalytic activity, in each case a reversible oxidative deamination reaction.

Isocitrate dehydrogenase 1 (NADP+), soluble is an enzyme that in humans is encoded by the IDH1 gene on chromosome 2. Isocitrate dehydrogenases catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. These enzymes belong to two distinct subclasses, one of which uses NAD+ as the electron acceptor and the other NADP+. Five isocitrate dehydrogenases have been reported: three NAD+-dependent isocitrate dehydrogenases, which localize to the mitochondrial matrix, and two NADP+-dependent isocitrate dehydrogenases, one of which is mitochondrial and the other predominantly cytosolic. Each NADP+-dependent isozyme is a homodimer. The protein encoded by this gene is the NADP+-dependent isocitrate dehydrogenase found in the cytoplasm and peroxisomes. It contains the PTS-1 peroxisomal targeting signal sequence. The presence of this enzyme in peroxisomes suggests roles in the regeneration of NADPH for intraperoxisomal reductions, such as the conversion of 2,4-dienoyl-CoAs to 3-enoyl-CoAs, as well as in peroxisomal reactions that consume 2-oxoglutarate, namely the alpha-hydroxylation of phytanic acid. The cytoplasmic enzyme serves a significant role in cytoplasmic NADPH production. Alternatively spliced transcript variants encoding the same protein have been found for this gene. [provided by RefSeq, Sep 2013]















