Related Research Articles

An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the ones which cause the common cold or influenza; drugs which inhibit growth of viruses are termed antiviral drugs or antivirals rather than antibiotics. They are also not effective against fungi; drugs which inhibit growth of fungi are called antifungal drugs.

Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance to the point that one or more drugs used to fight them are no longer effective. Fungi evolve antifungal resistance, viruses evolve antiviral resistance, protozoa evolve antiprotozoal resistance, and bacteria evolve antibiotic resistance. Together all of these come under the umbrella of antimicrobial resistance.

Drug resistance is the reduction in effectiveness of a medication such as an antimicrobial or an antineoplastic in treating a disease or condition. The term is used in the context of resistance that pathogens or cancers have "acquired", that is, resistance has evolved. Antimicrobial resistance and antineoplastic resistance challenge clinical care and drive research. When an organism is resistant to more than one drug, it is said to be multidrug-resistant.

Streptomycin is an antibiotic medication used to treat a number of bacterial infections, including tuberculosis, Mycobacterium avium complex, endocarditis, brucellosis, Burkholderia infection, plague, tularemia, and rat bite fever. For active tuberculosis it is often given together with isoniazid, rifampicin, and pyrazinamide. It is administered by injection into a vein or muscle.

Linezolid is an antibiotic used for the treatment of infections caused by Gram-positive bacteria that are resistant to other antibiotics. Linezolid is active against most Gram-positive bacteria that cause disease, including streptococci, vancomycin-resistant enterococci (VRE), and methicillin-resistant Staphylococcus aureus (MRSA). The main uses are infections of the skin and pneumonia although it may be used for a variety of other infections including drug-resistant tuberculosis. It is used either by injection into a vein or by mouth.
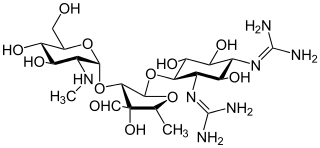
Aminoglycoside is a medicinal and bacteriologic category of traditional Gram-negative antibacterial medications that inhibit protein synthesis and contain as a portion of the molecule an amino-modified glycoside (sugar). The term can also refer more generally to any organic molecule that contains amino sugar substructures. Aminoglycoside antibiotics display bactericidal activity against Gram-negative aerobes and some anaerobic bacilli where resistance has not yet arisen but generally not against Gram-positive and anaerobic Gram-negative bacteria.
An antimicrobial is an agent that kills microorganisms (microbicide) or stops their growth. Antimicrobial medicines can be grouped according to the microorganisms they act primarily against. For example, antibiotics are used against bacteria, and antifungals are used against fungi. They can also be classified according to their function. The use of antimicrobial medicines to treat infection is known as antimicrobial chemotherapy, while the use of antimicrobial medicines to prevent infection is known as antimicrobial prophylaxis.

Vancomycin-resistant Staphylococcus aureus (VRSA) are strains of Staphylococcus aureus that have acquired resistance to the glycopeptide antibiotic vancomycin. Bacteria can acquire resistant genes either by random mutation or through the transfer of DNA from one bacterium to another. Resistance genes interfere with the normal antibiotic function and allow bacteria to grow in the presence of the antibiotic. Resistance in VRSA is conferred by the plasmid-mediated vanA gene and operon. Although VRSA infections are uncommon, VRSA is often resistant to other types of antibiotics and a potential threat to public health because treatment options are limited. VRSA is resistant to many of the standard drugs used to treat S. aureus infections. Furthermore, resistance can be transferred from one bacterium to another.
Multiple drug resistance (MDR), multidrug resistance or multiresistance is antimicrobial resistance shown by a species of microorganism to at least one antimicrobial drug in three or more antimicrobial categories. Antimicrobial categories are classifications of antimicrobial agents based on their mode of action and specific to target organisms. The MDR types most threatening to public health are MDR bacteria that resist multiple antibiotics; other types include MDR viruses, parasites.

Antibiotic sensitivity testing or antibiotic susceptibility testing is the measurement of the susceptibility of bacteria to antibiotics. It is used because bacteria may have resistance to some antibiotics. Sensitivity testing results can allow a clinician to change the choice of antibiotics from empiric therapy, which is when an antibiotic is selected based on clinical suspicion about the site of an infection and common causative bacteria, to directed therapy, in which the choice of antibiotic is based on knowledge of the organism and its sensitivities.

Lincosamides are a class of antibiotics, which include lincomycin, clindamycin, and pirlimycin.
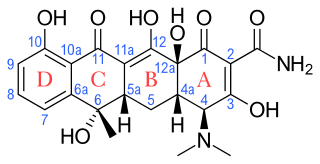
Tetracyclines are a group of broad-spectrum antibiotic compounds that have a common basic structure and are either isolated directly from several species of Streptomyces bacteria or produced semi-synthetically from those isolated compounds. Tetracycline molecules comprise a linear fused tetracyclic nucleus to which a variety of functional groups are attached. Tetracyclines are named after their four ("tetra-") hydrocarbon rings ("-cycl-") derivation ("-ine"). They are defined as a subclass of polyketides, having an octahydrotetracene-2-carboxamide skeleton and are known as derivatives of polycyclic naphthacene carboxamide. While all tetracyclines have a common structure, they differ from each other by the presence of chloro, methyl, and hydroxyl groups. These modifications do not change their broad antibacterial activity, but do affect pharmacological properties such as half-life and binding to proteins in serum.

Beta-lactamases are a family of enzymes involved in bacterial resistance to beta-lactam antibiotics. In bacterial resistance to beta-lactam antibiotics, the bacteria have beta-lactamase which degrade the beta-lactam rings, rendering the antibiotic ineffective. However, with beta-lactamase inhibitors, these enzymes on the bacteria are inhibited, thus allowing the antibiotic to take effect. Strategies for combating this form of resistance have included the development of new beta-lactam antibiotics that are more resistant to cleavage and the development of the class of enzyme inhibitors called beta-lactamase inhibitors. Although β-lactamase inhibitors have little antibiotic activity of their own, they prevent bacterial degradation of beta-lactam antibiotics and thus extend the range of bacteria the drugs are effective against.
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.
Persister cells are subpopulations of cells that resist treatment, and become antimicrobial tolerant by changing to a state of dormancy or quiescence. Persister cells in their dormancy do not divide. The tolerance shown in persister cells differs from antimicrobial resistance in that the tolerance is not inherited and is reversible. When treatment has stopped the state of dormancy can be reversed and the cells can reactivate and multiply. Most persister cells are bacterial, and there are also fungal persister cells, yeast persister cells, and cancer persister cells that show tolerance for cancer drugs.

Antibiotic use in livestock is the use of antibiotics for any purpose in the husbandry of livestock, which includes treatment when ill (therapeutic), treatment of a group of animals when at least one is diagnosed with clinical infection (metaphylaxis), and preventative treatment (prophylaxis). Antibiotics are an important tool to treat animal as well as human disease, safeguard animal health and welfare, and support food safety. However, used irresponsibly, this may lead to antibiotic resistance which may impact human, animal and environmental health.
The Society of Infectious Diseases Pharmacists (SIDP) is a non-profit organization comprising pharmacists and other allied health professionals specializing in infectious diseases and antimicrobial stewardship. According to the Board of Pharmaceutical Specialties, clinical pharmacists specializing in infectious diseases are trained in microbiology and pharmacology to develop, implement, and monitor drug regimens. These regimens incorporate the pharmacodynamics and pharmacokinetics of antimicrobials for patients.

Abigail A. Salyers was a microbiologist who pioneered the field of human microbiome research. Her work on the bacterial phylum Bacteroidetes and its ecology led to a better understanding of antibiotic resistance and mobile genetic elements. At a time where the prevailing paradigm was focused on E. coli as a model organism, Salyers emphasized the importance of investigating the breadth of microbial diversity. She was one of the first to conceptualize the human body as a microbial ecosystem. Over the course of her 40-year career, she was presented with numerous awards for teaching and research and an honorary degree from ETH Zurich, and served as president of the American Society for Microbiology.

Cresomycin is an experimental antibiotic. It binds to the bacterial ribosome in both Gram-negative and Gram-positive bacteria, and it has been found to be effective against multi-drug-resistant stains of Staphylococcus aureus, Escherichia coli, and Pseudomonas aeruginosa. It belongs to the bridged macrobicyclic oxepanoprolinamide antibiotics, which have similarities with lincosamides antibiotics.
References
- ↑ Davies, Julian E. (1964). "Studies on the Ribosomes of Streptomycin-Sensitive and Resistant Strains of Escherichia coli". Proceedings of the National Academy of Sciences. 51 (4): 659–664. Bibcode:1964PNAS...51..659D. doi: 10.1073/pnas.51.4.659 . ISSN 0027-8424. PMC 300136 . PMID 14166773.
- ↑ Davies, Julian; Jacob, François (28 September 1968). "Genetic mapping of the regulator and operator genes of the lac operon". Journal of Molecular Biology. 36 (3): 413–417. doi:10.1016/0022-2836(68)90165-4. ISSN 0022-2836. PMID 4939632.
- ↑ Yamada, Takeshi; Tipper, Donald; Davies, Julian (1968). "Enzymatic Inactivation of Streptomycin by R Factor-resistant Escherichia coli". Nature. 219 (5151): 288–291. Bibcode:1968Natur.219..288Y. doi:10.1038/219288a0. ISSN 0028-0836. PMID 4299554. S2CID 4172299.
- ↑ Davies, Julian (1974), King, Robert C. (ed.), "Bacterial Ribosomes", Bacteria, Bacteriophages, and Fungi: Volume 1, Boston, MA: Springer US, pp. 183–202, doi:10.1007/978-1-4899-1710-2_12, ISBN 978-1-4899-1710-2 , retrieved 11 September 2023
- ↑ Davies, J. (1 January 1979). "General Mechanisms of Antimicrobial Resistance". Clinical Infectious Diseases. 1 (1): 23–27. doi:10.1093/clinids/1.1.23. ISSN 1058-4838. PMID 400936.
- ↑ Davies, J.; Courvalin, P.; Berg, D. (1 January 1977). "Thoughts on the origins of resistance plasmids". Journal of Antimicrobial Chemotherapy. 3 (suppl C): 7–17. doi:10.1093/jac/3.suppl_c.7. ISSN 0305-7453. PMID 599132.
- ↑ Jimenez, Antonio; Davies, Julian (October 1980). "Expression of a transposable antibiotic resistance element in Saccharomyces". Nature. 287 (5785): 869–871. Bibcode:1980Natur.287..869J. doi:10.1038/287869a0. ISSN 0028-0836. PMID 6253817. S2CID 4368934.
- ↑ Davies, Julian (1 May 1973). "Bacteria Versus Antibiotics: Who Is Winning?". Annals of Internal Medicine. 78 (5): 813. doi:10.7326/0003-4819-78-5-813_6. ISSN 0003-4819.
- ↑ "Julian Davies". www.nasonline.org. Retrieved 6 September 2023.
- ↑ "Julian Davies". The Royal Society. Retrieved 30 April 2020.
- ↑ "Microbiology Society Prize Medallists". Microbiology Society. Retrieved 7 September 2023.