
The lactose operon is an operon required for the transport and metabolism of lactose in E. coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.

Inositol, or more precisely myo-inositol, is a carbocyclic sugar that is abundant in the brain and other mammalian tissues; it mediates cell signal transduction in response to a variety of hormones, neurotransmitters, and growth factors and participates in osmoregulation.
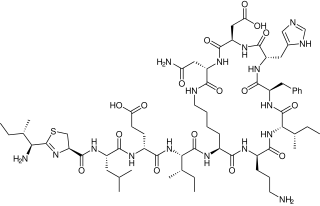
Bacitracin is a polypeptide antibiotic. It is a mixture of related cyclic peptides produced by Bacillus licheniformis bacteria, that was first isolated from the variety "Tracy I" in 1945. These peptides disrupt gram-positive bacteria by interfering with cell wall and peptidoglycan synthesis.

Catabolite activator protein is a trans-acting transcriptional activator that exists as a homodimer in solution. Each subunit of CAP is composed of a ligand-binding domain at the N-terminus and a DNA-binding domain at the C-terminus. Two cAMP molecules bind dimeric CAP with negative cooperativity. Cyclic AMP functions as an allosteric effector by increasing CAP's affinity for DNA. CAP binds a DNA region upstream from the DNA binding site of RNA Polymerase. CAP activates transcription through protein-protein interactions with the α-subunit of RNA Polymerase. This protein-protein interaction is responsible for (i) catalyzing the formation of the RNAP-promoter closed complex; and (ii) isomerization of the RNAP-promoter complex to the open conformation. CAP's interaction with RNA polymerase causes bending of the DNA near the transcription start site, thus effectively catalyzing the transcription initiation process. CAP's name is derived from its ability to affect transcription of genes involved in many catabolic pathways. For example, when the amount of glucose transported into the cell is low, a cascade of events results in the increase of cytosolic cAMP levels. This increase in cAMP levels is sensed by CAP, which goes on to activate the transcription of many other catabolic genes.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
The L-arabinose operon, also called the ara or araBAD operon, is an operon required for the breakdown of the five-carbon sugar L-arabinose in Escherichia coli. The L-arabinose operon contains three structural genes: araB, araA, araD, which encode for three metabolic enzymes that are required for the metabolism of L-arabinose. AraB (ribulokinase), AraA, AraD produced by these genes catalyse conversion of L-arabinose to an intermediate of the pentose phosphate pathway, D-xylulose-5-phosphate.

The Tryptophan operon leader is an RNA element found at the 5′ of some bacterial tryptophan operons. The leader sequence can form two different structures known as the terminator and the anti-terminator, based on the Tryptophan amounts in the cell. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon.

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.
The enzyme 5-dehydro-2-deoxyphosphogluconate aldolase catalyzes the chemical reaction
In enzymology, a 5-dehydro-2-deoxygluconokinase is an enzyme that catalyzes the chemical reaction
HutP is one of the anti-terminator proteins of Bacillus subtilis, which is responsible for regulating the expression of the hut structural genes of this organism in response to changes in the intracellular levels of L-histidine and divalent metal ions. In the hut operon, HutP is located just downstream from the promoter, while the five other subsequent structural genes, hutH, hutU, hutI, hutG and hutM, are positioned far downstream from the promoter. In the presence of L-histidine and divalent metal ions, HutP binds to the nascent hut mRNA leader transcript. This allows the anti-terminator to form, thereby preventing the formation of the terminator and permitting transcriptional read-through into the hut structural genes. In the absence of L-histidine and divalent metal ions or both, HutP does not bind to the hut mRNA, thus allowing the formation of a stem loop terminator structure within the nucleotide sequence located between the hutP and structural genes.
Carbon catabolite repression, or simply catabolite repression, is an important part of global control system of various bacteria and other microorganisms. Catabolite repression allows microorganisms to adapt quickly to a preferred carbon and energy source first. This is usually achieved through inhibition of synthesis of enzymes involved in catabolism of carbon sources other than the preferred one. The catabolite repression was first shown to be initiated by glucose and therefore sometimes referred to as the glucose effect. However, the term "glucose effect" is actually a misnomer since other carbon sources are known to induce catabolite repression.
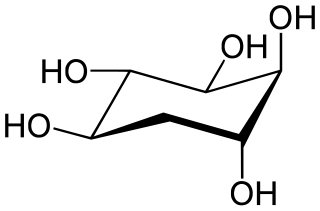
5-Deoxyinositol (quercitol) is a cyclitol. It can be found in wines aged in oak wood barrels. It can also be found in Quercus sp. (oaks) and in Gymnema sylvestre. It is different from quercetol, a synonym of quercetin.
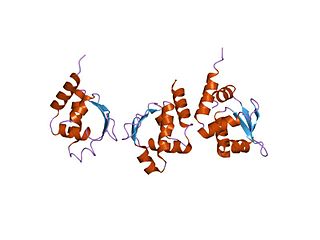
In molecular biology, the CodY protein family consists of several bacterial GTP-sensing transcriptional pleiotropic repressor CodY proteins. CodY has been found to repress the dipeptide transport operon (dpp) of Bacillus subtilis in nutrient-rich conditions. The CodY protein also has a repressor effect on many genes in Lactococcus lactis during growth in milk.
The Nucleobase cation symporter-2 (NCS2) family, also called the Nucleobase ascorbate transporter (NAT) family, consists of over 1000 sequenced proteins derived from gram-negative and gram-positive bacteria, archaea, fungi, plants and animals. The NCS2/NAT family is a member of the APC Superfamily of secondary carriers. Of the five known families of transporters that act on nucleobases, NCS2/NAT is the only one that is most widespread. Many functionally characterized members are specific for nucleobases including both purines and pyrimidines, but others are purine-specific. However, two closely related rat/human members of the family, SVCT1 and SVCT2, localized to different tissues of the body, co-transport L-ascorbate (vitamin C) and Na+ with a high degree of specificity and high affinity for the vitamin. Clustering of NCS2/NAT family members on the phylogenetic tree is complex, with bacterial proteins and eukaryotic proteins each falling into at least three distinct clusters. The plant and animal proteins cluster loosely together, but the fungal proteins branch from one of the three bacterial clusters forming a tighter grouping. E. coli possesses four distantly related paralogous members of the NCS2 family.
In molecular biology, the SR1 RNA is a small RNA (sRNA) produced by species of Bacillus and closely related bacteria. It is a dual-function RNA which acts both as a protein-coding RNA and as a regulatory sRNA.
Unsaturated rhamnogalacturonyl hydrolase (EC 3.2.1.172, YteR, YesR) is an enzyme with systematic name 2-O-(4-deoxy-beta-L-threo-hex-4-enopyranuronosyl)-alpha-L-rhamnopyranose hydrolase. This enzyme catalyses the following chemical reaction
The enzyme Rhamnogalacturonan endolyase is an enzyme with systematic name α-L-rhamnopyranosyl-(1→4)-α-D-galactopyranosyluronate endolyase. catalyses the following process:
In bacterial genetics, the mal regulon is a regulon - or group of genes under common regulation - associated with the catabolism of maltose and maltodextrins. The system is especially well characterized in the model organism Escherichia coli, where it is classically described as a group of ten genes in multiple operons whose expression is regulated by a single regulatory protein, malT. MalT binds to maltose or maltodextrin and undergoes a conformational change that allows it to bind DNA at sequences near the promoters of genes required for uptake and catabolism of these sugars. The maltose regulation system in E. coli is a classic example of positive regulation. malT is regulated by catabolite repression via the catabolite activator protein. Genes under the control of malT include ATP-binding cassette transporter components, maltoporin, maltose binding protein, and several enzymes. Other Gram-negative bacteria such as Klebsiella pneumoniae have additional genes under the control of malT.
Sulfoglycolysis is a catabolic process in primary metabolism in which sulfoquinovose (6-deoxy-6-sulfonato-glucose) is metabolized to produce energy and carbon-building blocks. Sulfoglycolysis pathways occur in a wide variety of organisms, and enable key steps in the degradation of sulfoquinovosyl diacylglycerol (SQDG), a sulfolipid found in plants and cyanobacteria into sulfite and sulfate. Sulfoglycolysis converts sulfoquinovose (C6H12O8S−) into various smaller metabolizable carbon fragments such as pyruvate and dihydroxyacetone phosphate that enter central metabolism. The free energy is used to form the high-energy molecules ATP (adenosine triphosphate) and NADH (reduced nicotinamide adenine dinucleotide). Unlike glycolysis, which allows metabolism of all carbons in glucose, some sulfoglycolysis pathways convert only a fraction of the carbon content of sulfoquinovose into smaller metabolizable fragments; the remaineder is excreted as C3-sulfonates 2,3-dihydroxypropanesulfonate (DHPS) or sulfolactate (SL); or the C2-sulfonate isethionate.









