A protein phosphatase is a phosphatase enzyme that removes a phosphate group from the phosphorylated amino acid residue of its substrate protein. Protein phosphorylation is one of the most common forms of reversible protein posttranslational modification (PTM), with up to 30% of all proteins being phosphorylated at any given time. Protein kinases (PKs) are the effectors of phosphorylation and catalyse the transfer of a γ-phosphate from ATP to specific amino acids on proteins. Several hundred PKs exist in mammals and are classified into distinct super-families. Proteins are phosphorylated predominantly on Ser, Thr and Tyr residues, which account for 79.3, 16.9 and 3.8% respectively of the phosphoproteome, at least in mammals. In contrast, protein phosphatases (PPs) are the primary effectors of dephosphorylation and can be grouped into three main classes based on sequence, structure and catalytic function. The largest class of PPs is the phosphoprotein phosphatase (PPP) family comprising PP1, PP2A, PP2B, PP4, PP5, PP6 and PP7, and the protein phosphatase Mg2+- or Mn2+-dependent (PPM) family, composed primarily of PP2C. The protein Tyr phosphatase (PTP) super-family forms the second group, and the aspartate-based protein phosphatases the third. The protein pseudophosphatases form part of the larger phosphatase family, and in most cases are thought to be catalytically inert, instead functioning as phosphate-binding proteins, integrators of signalling or subcellular traps. Examples of membrane-spanning protein phosphatases containing both active (phosphatase) and inactive (pseudophosphatase) domains linked in tandem are known, conceptually similar to the kinase and pseudokinase domain polypeptide structure of the JAK pseudokinases. A complete comparative analysis of human phosphatases and pseudophosphatases has been completed by Manning and colleagues, forming a companion piece to the ground-breaking analysis of the human kinome, which encodes the complete set of ~536 human protein kinases.

In biochemistry, a phosphatase is an enzyme that uses water to cleave a phosphoric acid monoester into a phosphate ion and an alcohol. Because a phosphatase enzyme catalyzes the hydrolysis of its substrate, it is a subcategory of hydrolases. Phosphatase enzymes are essential to many biological functions, because phosphorylation and dephosphorylation serve diverse roles in cellular regulation and signaling. Whereas phosphatases remove phosphate groups from molecules, kinases catalyze the transfer of phosphate groups to molecules from ATP. Together, kinases and phosphatases direct a form of post-translational modification that is essential to the cell's regulatory network.

The enzyme alkaline phosphatase has the physiological role of dephosphorylating compounds. The enzyme is found across a multitude of organisms, prokaryotes and eukaryotes alike, with the same general function but in different structural forms suitable to the environment they function in. Alkaline phosphatase is found in the periplasmic space of E. coli bacteria. This enzyme is heat stable and has its maximum activity at high pH. In humans, it is found in many forms depending on its origin within the body – it plays an integral role in metabolism within the liver and development within the skeleton. Due to its widespread prevalence in these areas, its concentration in the bloodstream is used by diagnosticians as a biomarker in helping determine diagnoses such as hepatitis or osteomalacia.
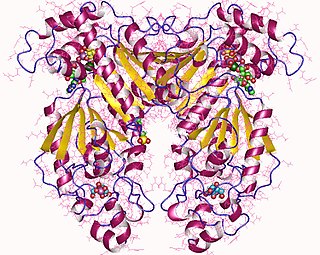
Phosphofructokinase-2 (6-phosphofructo-2-kinase, PFK-2) or fructose bisphosphatase-2 (FBPase-2), is an enzyme indirectly responsible for regulating the rates of glycolysis and gluconeogenesis in cells. It catalyzes formation and degradation of a significant allosteric regulator, fructose-2,6-bisphosphate (Fru-2,6-P2) from substrate fructose-6-phosphate. Fru-2,6-P2 contributes to the rate-determining step of glycolysis as it activates enzyme phosphofructokinase 1 in the glycolysis pathway, and inhibits fructose-1,6-bisphosphatase 1 in gluconeogenesis. Since Fru-2,6-P2 differentially regulates glycolysis and gluconeogenesis, it can act as a key signal to switch between the opposing pathways. Because PFK-2 produces Fru-2,6-P2 in response to hormonal signaling, metabolism can be more sensitively and efficiently controlled to align with the organism's glycolytic needs. This enzyme participates in fructose and mannose metabolism. The enzyme is important in the regulation of hepatic carbohydrate metabolism and is found in greatest quantities in the liver, kidney and heart. In mammals, several genes often encode different isoforms, each of which differs in its tissue distribution and enzymatic activity. The family described here bears a resemblance to the ATP-driven phospho-fructokinases, however, they share little sequence similarity, although a few residues seem key to their interaction with fructose 6-phosphate.

Protein tyrosine phosphatases (EC 3.1.3.48, systematic name protein-tyrosine-phosphate phosphohydrolase) are a group of enzymes that remove phosphate groups from phosphorylated tyrosine residues on proteins:
A nucleotidase is a hydrolytic enzyme that catalyzes the hydrolysis of a nucleotide into a nucleoside and a phosphate.
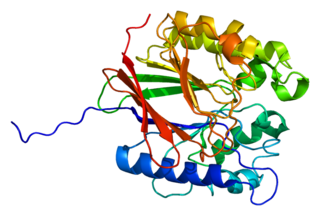
Tartrate-resistant acid phosphatase, also called acid phosphatase 5, tartrate resistant (ACP5), is a glycosylated monomeric metalloprotein enzyme expressed in mammals. It has a molecular weight of approximately 35kDa, a basic isoelectric point (7.6–9.5), and optimal activity in acidic conditions. TRAP is synthesized as latent proenzyme and activated by proteolytic cleavage and reduction. It is differentiated from other mammalian acid phosphatases by its resistance to inhibition by tartrate and by its molecular weight.
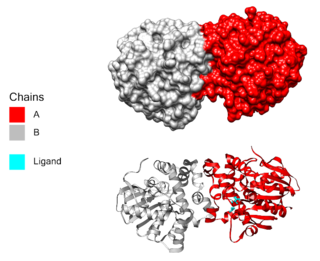
In enzymology, a haloacetate dehalogenase (EC 3.8.1.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a (R)-2-haloacid dehalogenase(EC 3.8.1.9), DL-2-haloacid halidohydrolase (inversion of configuration), DL-DEXi, (R,S)-2-haloacid dehalogenase (configuration-inverting)) is an enzyme that catalyzes the chemical reaction
In enzymology, a (S)-2-haloacid dehalogenase (EC 3.8.1.2) is an enzyme that catalyzes the chemical reaction
In enzymology, an undecaprenyl-diphosphatase (EC 3.6.1.27) is an enzyme that catalyzes the chemical reaction

The enzyme Inositol phosphate-phosphatase is of the phosphodiesterase family of enzymes. It is involved in the phosphophatidylinositol signaling pathway, which affects a wide array of cell functions, including but not limited to, cell growth, apoptosis, secretion, and information processing. Inhibition of inositol monophosphatase may be key in the action of lithium in treating bipolar disorder, specifically manic depression.
The enzyme phosphorylase a phosphatase (EC 3.1.3.17) catalyzes the reaction
The enzyme polynucleotide 5′-phosphatase (RNA 5′-triphosphatase, RTPase, EC 3.1.3.33) is an enzyme that catalyzes the reaction
In enzymology, a [isocitrate dehydrogenase (NADP+)] kinase (EC 2.7.11.5) is an enzyme that catalyzes the chemical reaction:

The haloacid dehydrogenase superfamily is a superfamily of enzymes that include phosphatases, phosphonatases, P-type ATPases, beta-phosphoglucomutases, phosphomannomutases, and dehalogenases, and are involved in a variety of cellular processes ranging from amino acid biosynthesis to detoxification.
Phospholipase C (EC 3.1.4.3, lipophosphodiesterase I, Clostridium welchii α-toxin, Clostridium oedematiens β- and γ-toxins, lipophosphodiesterase C, phosphatidase C, heat-labile hemolysin, α-toxin) is an enzyme with systematic name phosphatidylcholine cholinephosphohydrolase. This enzyme catalyses the following chemical reaction
8-oxo-dGDP phosphatase (EC 3.6.1.58, NUDT5) is an enzyme with systematic name 8-oxo-dGDP phosphohydrolase. This enzyme catalyses the following chemical reaction
2-haloacid dehalogenase (configuration-inverting) (EC 3.8.1.10, 2-haloalkanoic acid dehalogenase, 2-haloalkanoid acid halidohydrolase, DL-2-haloacid dehalogenase, DL-2-haloacid dehalogenase (inversion of configuration), DL-2-haloacid halidohydrolase (inversion of configuration), DL-DEXi, (R,S)-2-haloacid dehalogenase (configuration-inverting)) is an enzyme with systematic name (S)-2-haloacid dehalogenase (configuration-inverting). This enzyme catalyses the following chemical reaction
2-haloacid dehalogenase (configuration-retaining) (EC 3.8.1.11, 2-haloalkanoic acid dehalogenase, 2-haloalkanoid acid halidohydrolase, DL-2-haloacid dehalogenase, DL-DEXr) is an enzyme with systematic name (S)-2-haloacid dehalogenase (configuration-retaining). This enzyme catalyses the following chemical reaction








