This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.

An oncogene is a gene that has the potential to cause cancer. In tumor cells, they are often mutated or expressed at high levels.
In the context of gene regulation: transactivation is the increased rate of gene expression triggered either by biological processes or by artificial means, through the expression of an intermediate transactivator protein.

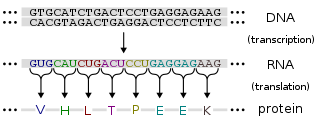
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
Functional genomics is a field of molecular biology that attempts to make use of the vast wealth of data given by genomic and transcriptomic projects to describe gene functions and interactions. Unlike structural genomics, functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. Functional genomics attempts to answer questions about the function of DNA at the levels of genes, RNA transcripts, and protein products. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional “gene-by-gene” approach.

In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking of expression is called repression.
Members of the signal transducer and activator of transcription (STAT) protein family are intracellular transcription factors that mediate many aspects of cellular immunity, proliferation, apoptosis and differentiation. They are primarily activated by membrane receptor-associated Janus kinases (JAK). Dysregulation of this pathway is frequently observed in primary tumors and leads to increased angiogenesis which enhances the survival of tumors and immunosuppression. Gene knockout studies have provided evidence that STAT proteins are involved in the development and function of the immune system and play a role in maintaining immune tolerance and tumor surveillance.
The classification of viral proteins as early proteins or late proteins depends on their relationship with genome replication. While many viruses are described as expressing early and late proteins, this definition of these terms is commonly reserved for class I DNA viruses.

Apetala 2 (AP2) is a gene and a member of a large family of transcription factors, the AP2/EREBP family. In Arabidopsis thaliana AP2 plays a role in the ABC model of flower development. It was originally thought that this family of proteins was plant-specific; however, recent studies have shown that apicomplexans, including the causative agent of malaria, Plasmodium falciparum encode a related set of transcription factors, called the ApiAP2 family.
CCAAT/enhancer-binding protein beta is a protein that in humans is encoded by the CEBPB gene.
Serine/threonine-protein kinases SGK represent a kinase subfamily with orthologs found across animal clades and in yeast. In most vertebrates, including humans, there are three isoforms encoded by the genes SGK1, SGK2, and SGK3. The name Serum/glucocorticoid-regulated kinase refers to the first cloning of a SGK family member from a cDNA library screen for genes upregulated by the glucocorticoid dexamethasone in a rat mammary epithelial tumor cell line.
The first human family member was cloned in a screen of hepatocellular genes regulated in response to cellular hydration or swelling.
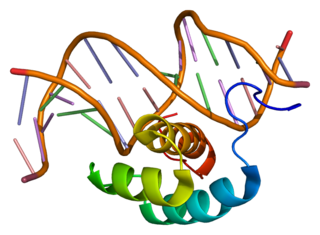
Homeobox protein Hox-B7 is a protein that in humans is encoded by the HOXB7 gene.

Homeobox protein Hox-B6 is a protein that in humans is encoded by the HOXB6 gene.

40S ribosomal protein S3a is a protein that in humans is encoded by the RPS3A gene.

Homeobox protein Hox-B4 is a protein that in humans is encoded by the HOXB4 gene.

Homeobox protein Hox-B2 is a protein that in humans is encoded by the HOXB2 gene.

ID4 is a protein coding gene. In humans, it encodes for the protein known as DNA-binding protein inhibitor ID-4.
This protein is known to be involved in the regulation of many cellular processes during both prenatal development and tumorigenesis. This is inclusive of embryonic cellular growth, senescence, cellular differentiation, apoptosis, and as an oncogene in angiogenesis.

LIM homeobox transcription factor 1, alpha, also known as LMX1A, is a protein which in humans is encoded by the LMX1A gene.

High mobility group nucleosome-binding domain-containing protein 3 is a protein that in humans is encoded by the HMGN3 gene.