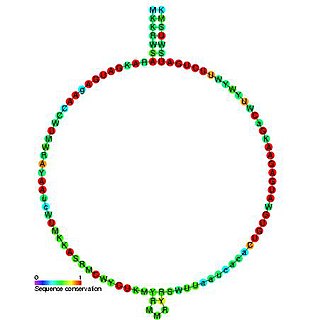
In molecular biology, Small nucleolar RNA Z110 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z110 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs Plant snoRNA Z110 was identified in screens of Arabidopsis thaliana and Oryza sativa .
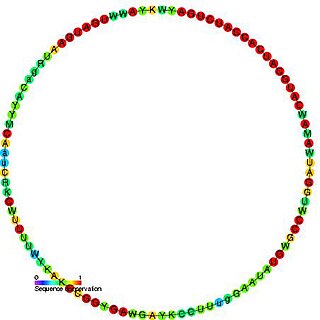
In molecular biology, Small nucleolar RNA snR55/Z10 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.

In molecular biology, Small nucleolar RNA snR52 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z13 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs.

In molecular biology, Small nucleolar RNA R12 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
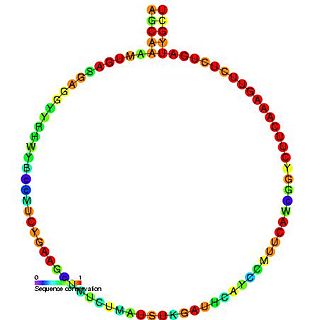
In molecular biology, Small nucleolar RNA R160 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA R66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
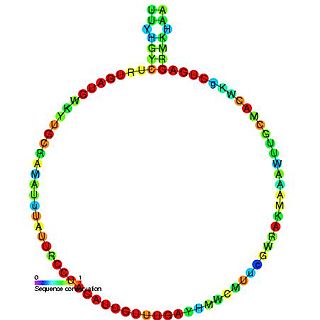
In molecular biology, snoRNA U34 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, the snoRNA snR48 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
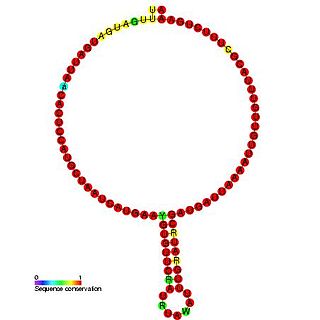
In molecular biology,snoRNA snR53 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR53 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR53 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
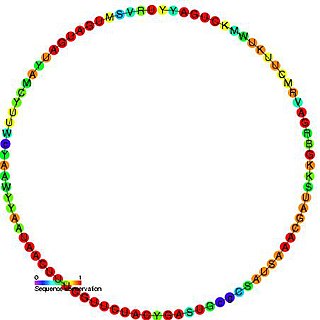
In molecular biology, snR54 is a non-coding RNA that is a member of the C/D class of snoRNA which contain the C box motif (UGAUGA) and D box motif (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. This snoRNA was first identified by a computational screen followed by experimental verification. This RNA guides the 2'-O-methylation of 18S rRNA. In yeast this snoRNA is found to reside in an intron of the IMD4 gene.
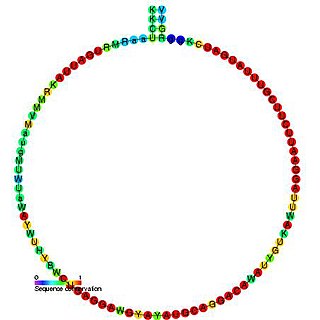
In molecular biology, snoRNA snR57 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR57 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR57 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
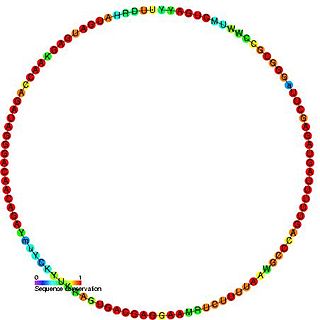
In molecular biology, snR64 is an RNA molecule belonging to the C/D class of small nucleolar RNA (snoRNA), which contain the C (UGAUGA) and D (CUGA) box motifs. Similar to most members of the box C/D family, snR64 is conjectured to help direct site-specific 2'-O-methylation of substrate RNAs.
snoRNA snR66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR66 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR66 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snoRNA snR69 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR69 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR69 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snoRNA snR71 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR71 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR71 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.

In molecular biology, Small nucleolar RNA Z122 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA Z194 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, Small nucleolar RNA Z221 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
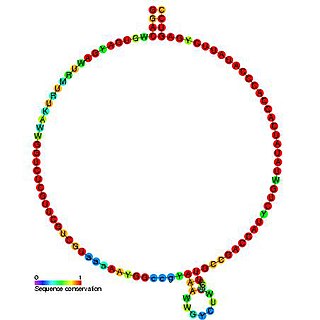
In molecular biology, Small nucleolar RNA Z43 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z43 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z43 was identified in a screen of Oryza sativa.
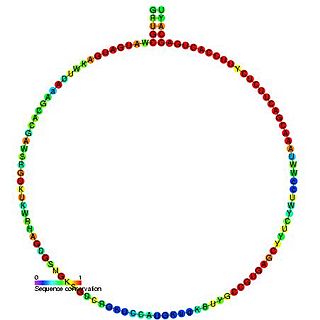
In molecular biology, Small nucleolar RNA Z152 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z152 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z152 was identified in screens of Oryza sativa and Arabidopsis thaliana.




















