Related Research Articles

G protein-coupled receptors (GPCRs), also known as seven-(pass)-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptors, and G protein-linked receptors (GPLR), form a large group of evolutionarily-related proteins that are cell surface receptors that detect molecules outside the cell and activate cellular responses. Coupling with G proteins, they are called seven-transmembrane receptors because they pass through the cell membrane seven times. Ligands can bind either to extracellular N-terminus and loops or to the binding site within transmembrane helices. They are all activated by agonists although a spontaneous auto-activation of an empty receptor can also be observed.

G proteins, also known as guanine nucleotide-binding proteins, are a family of proteins that act as molecular switches inside cells, and are involved in transmitting signals from a variety of stimuli outside a cell to its interior. Their activity is regulated by factors that control their ability to bind to and hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP). When they are bound to GTP, they are 'on', and, when they are bound to GDP, they are 'off'. G proteins belong to the larger group of enzymes called GTPases.

Integrins are transmembrane receptors that facilitate cell-extracellular matrix (ECM) adhesion. Upon ligand binding, integrins activate signal transduction pathways that mediate cellular signals such as regulation of the cell cycle, organization of the intracellular cytoskeleton, and movement of new receptors to the cell membrane. The presence of integrins allows rapid and flexible responses to events at the cell surface.

Pharmacology is a branch of medicine and pharmaceutical sciences which is concerned with the study of drug or medication action, where a drug can be broadly or narrowly defined as any man-made, natural, or endogenous molecule which exerts a biochemical or physiological effect on the cell, tissue, organ, or organism. More specifically, it is the study of the interactions that occur between a living organism and chemicals that affect normal or abnormal biochemical function. If substances have medicinal properties, they are considered pharmaceuticals.

A protein kinase is a kinase which selectively modifies other proteins by covalently adding phosphates to them (phosphorylation) as opposed to kinases which modify lipids, carbohydrates, or other molecules. Phosphorylation usually results in a functional change of the target protein (substrate) by changing enzyme activity, cellular location, or association with other proteins. The human genome contains about 500 protein kinase genes and they constitute about 2% of all human genes. There are two main types of protein kinase, the great majority are serine/threonine kinases, which phosphorylate the hydroxyl groups of serines and threonines in their targets and the other are tyrosine kinases, although additional types exist. Protein kinases are also found in bacteria and plants. Up to 30% of all human proteins may be modified by kinase activity, and kinases are known to regulate the majority of cellular pathways, especially those involved in signal transduction.

Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.

Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molecules, biopharmaceuticals including peptides and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.

Pharmacodynamics (PD) is the study of the biochemical and physiologic effects of drugs. The effects can include those manifested within animals, microorganisms, or combinations of organisms.
A biological target is anything within a living organism to which some other entity is directed and/or binds, resulting in a change in its behavior or function. Examples of common classes of biological targets are proteins and nucleic acids. The definition is context-dependent, and can refer to the biological target of a pharmacologically active drug compound, the receptor target of a hormone, or some other target of an external stimulus. Biological targets are most commonly proteins such as enzymes, ion channels, and receptors.
Neuropharmacology is the study of how drugs affect cellular function in the nervous system, and the neural mechanisms through which they influence behavior. There are two main branches of neuropharmacology: behavioral and molecular. Behavioral neuropharmacology focuses on the study of how drugs affect human behavior (neuropsychopharmacology), including the study of how drug dependence and addiction affect the human brain. Molecular neuropharmacology involves the study of neurons and their neurochemical interactions, with the overall goal of developing drugs that have beneficial effects on neurological function. Both of these fields are closely connected, since both are concerned with the interactions of neurotransmitters, neuropeptides, neurohormones, neuromodulators, enzymes, second messengers, co-transporters, ion channels, and receptor proteins in the central and peripheral nervous systems. Studying these interactions, researchers are developing drugs to treat many different neurological disorders, including pain, neurodegenerative diseases such as Parkinson's disease and Alzheimer's disease, psychological disorders, addiction, and many others.
A biochemical cascade, also known as a signaling cascade or signaling pathway, is a series of chemical reactions that occur within a biological cell when initiated by a stimulus. This stimulus, known as a first messenger, acts on a receptor that is transduced to the cell interior through second messengers which amplify the signal and transfer it to effector molecules, causing the cell to respond to the initial stimulus. Most biochemical cascades are series of events, in which one event triggers the next, in a linear fashion. At each step of the signaling cascade, various controlling factors are involved to regulate cellular actions, in order to respond effectively to cues about their changing internal and external environments.

Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.

KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.
In biology, cell signaling or cell-cell communication, governs the basic activities of cells and coordinates multiple-cell actions. A signal is an entity that codes or conveys information. Biological processes are complex molecular interactions that involve a lot of signals. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity, as well as normal tissue homeostasis. Errors in signaling interactions and cellular information processing may cause diseases such as cancer, autoimmunity, and diabetes. By understanding cell signaling, clinicians may treat diseases more effectively and, theoretically, researchers may develop artificial tissues.
Stuart L. Schreiber is a scientist at Harvard University and co-Founder of the Broad Institute. He has been active in chemical biology, especially the use of small molecules as probes of biology and medicine. Small molecules are the molecules of life most associated with dynamic information flow; these work in concert with the macromolecules that are the basis for inherited information flow.

P2Y receptors are a family of purinergic G protein-coupled receptors, stimulated by nucleotides such as ATP, ADP, UTP, UDP and UDP-glucose. To date, 8 P2Y receptors have been cloned in humans: P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13 and P2Y14.
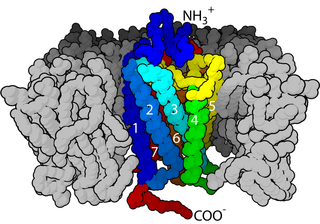
Cell surface receptors are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the extracellular space. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane.

György Kéri was a Hungarian biochemist, professor and Doctor of Biological Sciences (D.Sc.). His major field of research was signal transduction therapy and he participated in the development of novel drug discovery technologies and drug candidates that entered the clinical development process.
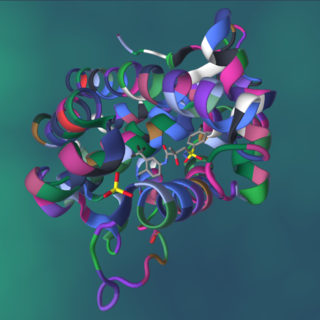
A protein–ligand complex is a complex of a protein bound with a ligand that is formed following molecular recognition between proteins that interact with each other or with various other molecules. Formation of a protein-ligand complex is based on molecular recognition between biological macromolecules and ligands, where ligand means any molecule that binds the protein with high affinity and specificity. Molecular recognition is not a process by itself since it is part of a functionally important mechanism involving the essential elements of life like in self-replication, metabolism, and information processing. For example DNA-replication depends on recognition and binding of DNA double helix by helicase, DNA single strand by DNA-polymerase and DNA segments by ligase. Molecular recognition depends on affinity and specificity. Specificity means that proteins distinguish the highly specific binding partner from less specific partners and affinity allows the specific partner with high affinity to remain bound even if there are high concentrations of less specific partners with lower affinity.
References
- Hanzlik RP, Koen YM, Theertham B, Dong Y, Fang J (2007). "The reactive metabolite target protein database (TPDB) – a web-accessible resource". BMC Bioinformatics. 8: 95. doi:10.1186/1471-2105-8-95. PMC 1832215 . PMID 17367530.