
An alpha helix is a sequence of amino acids in a protein that are twisted into a coil.

A ubiquitin ligase is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases regulates diverse areas such as cell trafficking, DNA repair, and signaling and is of profound importance in cell biology. E3 ligases are also key players in cell cycle control, mediating the degradation of cyclins, as well as cyclin dependent kinase inhibitor proteins. The human genome encodes over 600 putative E3 ligases, allowing for tremendous diversity in substrates.

Helix-turn-helix is a DNA-binding domain (DBD). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

F-box proteins are proteins containing at least one F-box domain. The first identified F-box protein is one of three components of the SCF complex, which mediates ubiquitination of proteins targeted for degradation by the 26S proteasome.
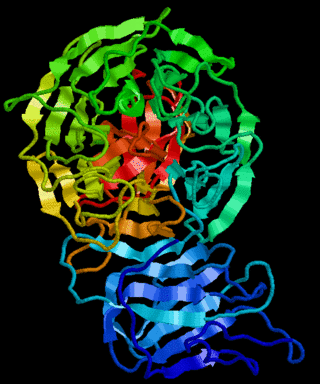
Kelch proteins are a widespread group of proteins that contain multiple Kelch motifs. The kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a β-propeller tertiary structure. Kelch-repeat β-propellers are generally involved in protein–protein interactions, though the large diversity of domain architectures and limited sequence identity between kelch motifs make characterisation of the kelch superfamily difficult.

Zinc finger and BTB domain-containing protein 16 is a protein that in humans is encoded by the ZBTB16 gene.

RING-box protein 2 is a protein that in humans is encoded by the RNF7 gene.

Zinc finger protein 238 is a zinc finger containing transcription factor that in humans is encoded by the ZNF238 gene.

In molecular biology, a RING (short for Really Interesting New Gene) finger domain is a protein structural domain of zinc finger type which contains a C3HC4 amino acid motif which binds two zinc cations (seven cysteines and one histidine arranged non-consecutively). This protein domain contains 40 to 60 amino acids. Many proteins containing a RING finger play a key role in the ubiquitination pathway. Conversely, proteins with RING finger domains are the largest type of ubiquitin ligases in the human genome.

Zinc finger and BTB domain-containing protein 32 is a protein that in humans is encoded by the 1960 bp ZBTB32 gene. The 52 kDa protein is a transcriptional repressor and the gene is expressed in T and B cells upon activation, but also significantly in testis cells. It is a member of the Poxviruses and Zinc-finger (POZ) and Krüppel (POK) family of proteins, and was identified in multiple screens involving either immune cell tumorigenesis or immune cell development.
The RhoBTB family is a subgroup of the Rho family of small GTPases. They are a highly divergent class and are all characterized by an N-terminal Rho-related domain followed by at least one C-terminal BTB domain.
In molecular biology the ZZ-type zinc finger domain is a type of protein domain that was named because of its ability to bind two zinc ions. These domains contain 4-6 Cys residues that participate in zinc binding, including a Cys-X2-Cys motif found in other zinc finger domains. These zinc fingers are thought to be involved in protein-protein interactions. The structure of the ZZ domain shows that it belongs to the family of cross-brace zinc finger motifs that include the PHD, RING, and FYVE domains. ZZ-type zinc finger domains are found in:

In molecular biology the B-box-type zinc finger domain is a short protein domain of around 40 amino acid residues in length. B-box zinc fingers can be divided into two groups, where types 1 and 2 B-box domains differ in their consensus sequence and in the spacing of the 7-8 zinc-binding residues. Several proteins contain both types 1 and 2 B-boxes, suggesting some level of cooperativity between these two domains.
In molecular biology, the BEN domain is a protein domain which is found in diverse proteins including:
In molecular biology, the FLYWCH zinc finger is a zinc finger domain. It is found in a number of eukaryotic proteins. FLYWCH is a C2H2-type zinc finger characterised by five conserved hydrophobic residues, containing the conserved sequence motif:
F/Y-X(n)-L-X(n)-F/Y-X(n)-WXCX(6-12)CX(17-22)HXH

The WRKY domain is found in the WRKY transcription factor family, a class of transcription factors. The WRKY domain is found almost exclusively in plants although WRKY genes appear present in some diplomonads, social amoebae and other amoebozoa, and fungi incertae sedis. They appear absent in other non-plant species. WRKY transcription factors have been a significant area of plant research for the past 20 years. The WRKY DNA-binding domain recognizes the W-box (T)TGAC(C/T) cis-regulatory element.
In molecular biology, this protein domain has been termed SRA-YDG, which is the abbreviation for SET and Ring finger Associated, YDG motif. Additional characteristics of the domain include conservation of up to 13 evenly spaced glycine residues and a VRV(I/V)RG motif. The protein domain is mainly found in plants and animals and in bacteria.

Intermediate filament family orphan 1 is a protein that in humans is encoded by the IFFO1 gene. IFFO1 has uncharacterized function and a weight of 61.98 kDa. IFFO1 proteins play an important role in the cytoskeleton and the nuclear envelope of most eukaryotic cell types.

Ubiquitin-binding domains (UBDs) are protein domains that recognise and bind non-covalently to ubiquitin through protein-protein interactions. As of 2019, a total of 29 types of UBDs had been identified in the human proteome. Most UBDs bind to ubiquitin only weakly, with binding affinities in the low to mid μM range. Proteins containing UBDs are known as ubiquitin-binding proteins or sometimes as "ubiquitin receptors".














