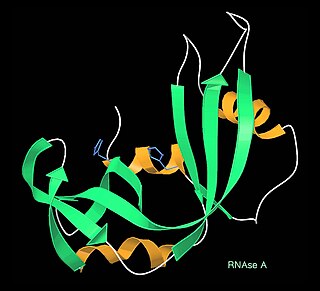
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations. The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB.
A kinemage is an interactive graphic scientific illustration. It often is used to visualize molecules, especially proteins although it can also represent other types of 3-dimensional data. The kinemage system is designed to optimize ease of use, interactive performance, and the perception and communication of detailed 3D information. The kinemage information is stored in a text file, human- and machine-readable, that describes the hierarchy of display objects and their properties, and includes optional explanatory text. The kinemage format is a defined chemical MIME type of 'chemical/x-kinemage' with the file extension '.kin'.
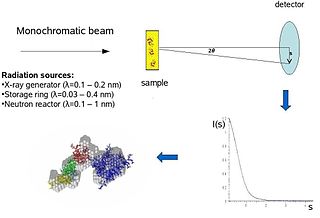
Biological small-angle scattering is a small-angle scattering method for structure analysis of biological materials. Small-angle scattering is used to study the structure of a variety of objects such as solutions of biological macromolecules, nanocomposites, alloys, and synthetic polymers. Small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS) are the two complementary techniques known jointly as small-angle scattering (SAS). SAS is an analogous method to X-ray and neutron diffraction, wide angle X-ray scattering, as well as to static light scattering. In contrast to other X-ray and neutron scattering methods, SAS yields information on the sizes and shapes of both crystalline and non-crystalline particles. When used to study biological materials, which are very often in aqueous solution, the scattering pattern is orientation averaged.
Nuclear magnetic resonance spectroscopy of proteins is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, and Angela Gronenborn at the NIH, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated.

PK-11195 is an isoquinoline carboxamide which binds selectively to the peripheral benzodiazepine receptor (PBR). It is one of the most commonly used PBR ligands due to its high affinity for the PBR in all species, although it is starting to be replaced by newer and more selective ligands.
Axel T. Brunger is a German American biophysicist. He is Professor of Molecular and Cellular Physiology, and Neurology, of Photon Science and, by courtesy, of Structural Biology at Stanford University, and a Howard Hughes Medical Institute Investigator. He also is currently serving as the Chair of the Department of Molecular and Cellular Physiology (2013–present).
Fragment-based lead discovery (FBLD) also known as fragment-based drug discovery (FBDD) is a method used for finding lead compounds as part of the drug discovery process. Fragments are small organic molecules which are small in size and low in molecular weight. It is based on identifying small chemical fragments, which may bind only weakly to the biological target, and then growing them or combining them to produce a lead with a higher affinity. FBLD can be compared with high-throughput screening (HTS). In HTS, libraries with up to millions of compounds, with molecular weights of around 500 Da, are screened, and nanomolar binding affinities are sought. In contrast, in the early phase of FBLD, libraries with a few thousand compounds with molecular weights of around 200 Da may be screened, and millimolar affinities can be considered useful. FBLD is a technique being used in research for discovering novel potent inhibitors.
Structure-Based Assignment (SBA) is a technique to accelerate the resonance assignment which is a key bottleneck of NMR structural biology. A homologous (similar) protein is used as a template to the target protein in SBA. This template protein provides prior structural information about the target protein and leads to faster resonance assignment. By analogy, in X-ray Crystallography, the molecular replacement technique allows solution of the crystallographic phase problem when a homologous structural model is known, thereby facilitating rapid structure determination. Some of the SBA algorithms are CAP which is an RNA assignment algorithm which performs an exhaustive search over all permutations, MARS which is a program for robust automatic backbone assignment and Nuclear Vector Replacement (NVR) which is a molecular replacement like approach for SBA of resonances and sparse Nuclear Overhauser Effect (NOE)'s.

Protein crystallization is the process of formation of a protein crystal. Protein crystals are useful in the study of protein structures for use in medicine, amongst other applications. In the process of protein crystallization, proteins are dissolved in an aqueous environment and sample solution in order to reach the supersaturated state. This supersaturated state allows researchers to study the internal structure of proteins. Different methods are used to reach that state such as vapor diffusion, microbatch, microdialysis, and free-interface diffusion. Developing protein crystals is difficult, as the process is influenced by many factors, including pH, temperature, ionic strength in the crystallization solution, and even gravity. Once properly developed, these crystals can be used in structural biology to study the molecular structure of the protein, particularly for various industrial or biotechnological purposes, such as developing cancer treatment.

Antony John Williams is a British chemist and expert in the fields of both nuclear magnetic resonance (NMR) spectroscopy and cheminformatics at the United States Environmental Protection Agency. He is the founder of the ChemSpider website that was purchased by the Royal Society of Chemistry in May 2009. He is a science blogger, one of the hosts of the SciMobileApps wiki, a community-based wiki for Scientific Mobile Apps and an author.
Nuclear magnetic resonance crystallography is a method which utilizes primarily NMR spectroscopy to determine the structure of solid materials on the atomic scale. Thus, solid-state NMR spectroscopy would be used primarily, possibly supplemented by quantum chemistry calculations, powder diffraction etc. If suitable crystals can be grown, any crystallographic method would generally be preferred to determine the crystal structure comprising in case of organic compounds the molecular structures and molecular packing. The main interest in NMR crystallography is in microcrystalline materials which are amenable to this method but not to X-ray, neutron and electron diffraction. This is largely because interactions of comparably short range are measured in NMR crystallography.
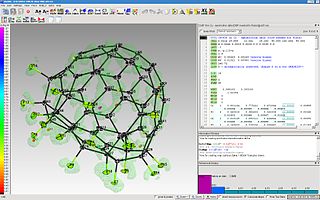
The program ShelXle is a graphical user interface for the structure refinement program SHELXL. ShelXle combines an editor with syntax highlighting for the SHELXL-associated .ins (input) and .res (output) files with an interactive graphical display for visualization of a three-dimensional structure including the electron density (Fo) and difference density (Fo-Fc) maps.
Biomolecular complex, also called macromolecular complex or biomacromolecular complex, is any biological complex made of more than one molecule of protein, RNA, DNA, lipids, or carbohydrates. The interactions between these biomolecules are non-covalent. Examples:

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
Protein chemical shift prediction is a branch of biomolecular nuclear magnetic resonance spectroscopy that aims to accurately calculate protein chemical shifts from protein coordinates. Protein chemical shift prediction was first attempted in the late 1960s using semi-empirical methods applied to protein structures solved by X-ray crystallography. Since that time protein chemical shift prediction has evolved to employ much more sophisticated approaches including quantum mechanics, machine learning and empirically derived chemical shift hypersurfaces. The most recently developed methods exhibit remarkable precision and accuracy.
Resolution by Proxy (ResProx) is a method for assessing the equivalent X-ray resolution of NMR-derived protein structures. ResProx calculates resolution from coordinate data rather than from electron density or other experimental inputs. This makes it possible to calculate the resolution of a structure regardless of how it was solved. ResProx was originally designed to serve as a simple, single-number evaluation that allows straightforward comparison between the quality/resolution of X-ray structures and the quality of a given NMR structure. However, it can also be used to assess the reliability of an experimentally reported X-ray structure resolution, to evaluate protein structures solved by unconventional or hybrid means and to identify fraudulent structures deposited in the PDB. ResProx incorporates more than 25 different structural features to determine a single resolution-like value. ResProx values are reported in Angstroms. Tests on thousands of X-ray structures show that ResProx values match very closely to resolution values reported by X-ray crystallographers. Resolution-by-proxy values can be calculated for newly determined protein structures using a freely accessible ResProx web server. This server accepts protein coordinate data and generates a resolution estimate for that input structure.

Randy John Read is a Wellcome Trust Principal Research Fellow and Professor of Protein Crystallography at the University of Cambridge.
The SBGrid Consortium is an innovative global research computing group financially supported by participating research laboratories and operated out of Harvard Medical School. SBGrid provides the global structural biology community with support for research computing. Members of the SBGrid Consortium fund SBGrid’s ongoing operations through an annual membership fee. The resulting organization is a user-supported and user-directed community resource.








