
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the BRCA1 gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. BRCA1 is a human tumor suppressor gene and is responsible for repairing DNA.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.

8-Oxoguanine glycosylase, also known as OGG1 is a DNA glycosylase enzyme that, in humans, is encoded by the OGG1 gene. It is involved in base excision repair. It is found in bacterial, archaeal and eukaryotic species.

The growth-hormone-releasing hormone receptor (GHRHR) is a G-protein-coupled receptor that binds growth hormone-releasing hormone. The GHRHR activates a Gs protein that causes a cascade of cAMP via adenylate cyclase.

Transcription activator BRG1 also known as ATP-dependent chromatin remodeler SMARCA4 is a protein that in humans is encoded by the SMARCA4 gene.

β-Tropomyosin, also known as tropomyosin beta chain is a protein that in humans is encoded by the TPM2 gene. β-tropomyosin is striated muscle-specific coiled coil dimer that functions to stabilize actin filaments and regulate muscle contraction.

RAD51 homolog C , also known as RAD51C, is a protein which in humans is encoded by the RAD51C gene.

TAR DNA-binding protein 43, is a protein that in humans is encoded by the TARDBP gene.

Homeobox protein Hox-C6 is a protein that in humans is encoded by the HOXC6 gene.

Homeobox protein Hox-C5 is a protein that in humans is encoded by the HOXC5 gene.

Lys-63-specific deubiquitinase BRCC36 is an enzyme that in humans is encoded by the BRCC3 gene.

FAD-dependent oxidoreductase domain-containing protein 1 (FOXRED1), also known as H17, or FP634 is an enzyme that in humans is encoded by the FOXRED1 gene. FOXRED1 is an oxidoreductase and complex I-specific molecular chaperone involved in the assembly and stabilization of NADH dehydrogenase (ubiquinone) also known as complex I, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in FOXRED1 have been associated with Leigh syndrome and infantile-onset mitochondrial encephalopathy.

Homeobox protein MOX-1 is a protein that in humans is encoded by the MEOX1 gene.
Periannan Senapathy is a molecular biologist, geneticist, author and entrepreneur. He is the founder, president and chief scientific officer at Genome International Corporation, a biotechnology, bioinformatics, and information technology firm based in Madison, Wisconsin, which develops computational genomics applications of next-generation DNA sequencing (NGS) and clinical decision support systems for analyzing patient genome data that aids in diagnosis and treatment of diseases.
Megf8 also known as Multiple Epidermal Growth Factor-like Domains 8, is a protein coding gene that encodes a single pass membrane protein, known to participate in developmental regulation and cellular communication. It is located on chromosome 19 at the 49th open reading frame in humans (19q13.2). There are two isoform constructs known for MEGF8, which differ by a 67 amino acid indel. The isoform 2 splice version is 2785 amino acids long, and predicted to be 296.6 kdal in mass. Isoform 1 is composed of 2845 amino acids and predicted to weigh 303.1 kdal. Using BLAST searches, orthologs were found primarily in mammals, but MEGF8 is also conserved in invertebrates and fishes, and rarely in birds, reptiles, and amphibians. A notably important paralog to multiple epidermal growth factor-like domains 8 is ATRNL1, which is also a single pass transmembrane protein, with several of the same key features and motifs as MEGF8, as indicated by Simple Modular Architecture Research Tool (SMART) which is hosted by the European Molecular Biology Laboratory located in Heidelberg, Germany. MEGF8 has been predicted to be a key player in several developmental processes, such as left-right patterning and limb formation. Currently, researchers have found MEGF8 SNP mutations to be the cause of Carpenter syndrome subtype 2.

A minigene is a minimal gene fragment that includes an exon and the control regions necessary for the gene to express itself in the same way as a wild type gene fragment. This is a minigene in its most basic sense. More complex minigenes can be constructed containing multiple exons and intron(s). Minigenes provide a valuable tool for researchers evaluating splicing patterns both in vivo and in vitro biochemically assessed experiments. Specifically, minigenes are used as splice reporter vectors and act as a probe to determine which factors are important in splicing outcomes. They can be constructed to test the way both cis-regulatory elements and trans-regulatory elements affect gene expression.
Single nucleotide polymorphism annotation is the process of predicting the effect or function of an individual SNP using SNP annotation tools. In SNP annotation the biological information is extracted, collected and displayed in a clear form amenable to query. SNP functional annotation is typically performed based on the available information on nucleic acid and protein sequences.
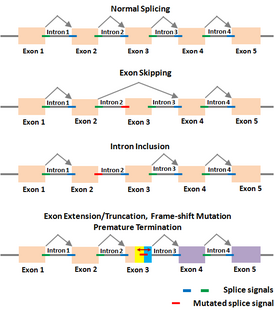
The Shapiro–Senapathy algorithm (S&S) is an algorithm for predicting the splice sites, exons and genes in animals and plants. This algorithm has the ability to discover disease-causing mutations in splice junctions in cancerous and non-cancerous diseases that is being used in major research institutions around the world.
David Scott Wishart is a researcher in the fields of metabolomics, cheminformatics and bioinformatics. He is a professor in the Department of Biological Sciences at the University of Alberta, with cross appointments in the Faculty of Pharmacy and Pharmaceutical Sciences and the Department of Laboratory Medicine and Pathology. He also holds a joint appointment in metabolomics at the Pacific Northwest National Laboratory in Richland, Washington.

Francisco Ernesto (Tito) Baralle is an Argentinian geneticist best known for his innovations in molecular biology and in particular the discovery of how genes are processed and mechanisms in mRNA splicing.















