
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed from an RNA. cDNA exists in both single-stranded and double-stranded forms and in both natural and engineered forms.
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word intron is derived from the term intragenic region, i.e., a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons.

A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backward). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.

Retrotransposons are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged.
Pseudoviridae is a family of viruses, which includes three genera.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
Simian foamy virus (SFV), historically Human foamy virus (HFV), is a species of the genus Spumavirus that belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including prosimians, New World and Old World monkeys, as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques, and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat-hunting practices.
Exon shuffling is a molecular mechanism for the formation of new genes. It is a process through which two or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron structure. There are different mechanisms through which exon shuffling occurs: transposon mediated exon shuffling, crossover during sexual recombination of parental genomes and illegitimate recombination.

Multicopy single-stranded DNA (msDNA) is a type of extrachromosomal satellite DNA that consists of a single-stranded DNA molecule covalently linked via a 2'-5'phosphodiester bond to an internal guanosine of an RNA molecule. The resultant DNA/RNA chimera possesses two stem-loops joined by a branch similar to the branches found in RNA splicing intermediates. The coding region for msDNA, called a "retron", also encodes a type of reverse transcriptase, which is essential for msDNA synthesis.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.
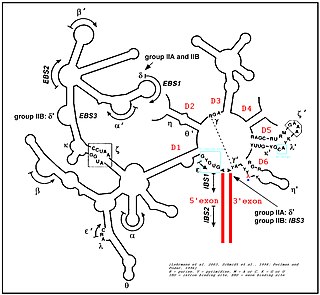
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically insert into DNA sites has been exploited as a tool for biotechnology. For example, group II introns can be modified to make site-specific genome insertions and deliver cargo DNA such as reporter genes or lox sites
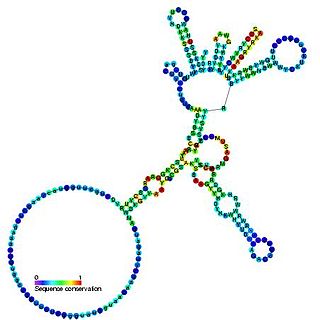
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

A retron is a distinct DNA sequence found in the genome of many bacteria species that codes for reverse transcriptase and a unique single-stranded DNA/RNA hybrid called multicopy single-stranded DNA (msDNA). Retron msr RNA is the non-coding RNA produced by retron elements and is the immediate precursor to the synthesis of msDNA. The retron msr RNA folds into a characteristic secondary structure that contains a conserved guanosine residue at the end of a stem loop. Synthesis of DNA by the retron-encoded reverse transcriptase (RT) results in a DNA/RNA chimera which is composed of small single-stranded DNA linked to small single-stranded RNA. The RNA strand is joined to the 5′ end of the DNA chain via a 2′–5′ phosphodiester linkage that occurs from the 2′ position of the conserved internal guanosine residue.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.

Maturase K (matK) is a plant plastidial gene. The protein it encodes is an organelle intron maturase, a protein that splices Group II introns. It is essential for in vivo splicing of Group II introns. Amongst other maturases, this protein retains only a well conserved domain X and remnants of a reverse transcriptase domain.
Semotivirus is the only genus of viruses in the family Belpaoviridae. Species exist as retrotransposons in a eukaryotic host's genome. BEL/pao transposons are only found in animals. Semotivirus is the only genus currently recognized, the genus description corresponds to the family, Belpaoviridae description.

Ortervirales is an order that contains all accepted species of single-stranded RNA viruses that replicate through a DNA intermediate and all accepted species of double-stranded DNA viruses that replicate through an RNA intermediate . The name is derived from the reverse of retro.

Alan Lambowitz is a professor for the University of Texas at Austin in Molecular Biosciences and Oncology and has been instrumental in many bio-molecular processes and concepts, such as intron splicing and mitochondrial ribosomal assembly.
Ravindra N. Singh is an Indian American scientist, inventor and academic. He is a professor in the Department of Biomedical Sciences of the College of Veterinary Medicine at Iowa State University.










