
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane. Peripheral membrane proteins are transiently associated with the cell membrane.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
A membrane transport protein is a membrane protein involved in the movement of ions, small molecules, and macromolecules, such as another protein, across a biological membrane. Transport proteins are integral transmembrane protein; that is they exist permanently within and span the membrane across which they transport substances. The proteins may assist in the movement of substances by facilitated diffusion or active transport. The two main types of proteins involved in such transport are broadly categorized as either channels or carriers. The solute carriers and atypical SLCs are secondary active or facilitative transporters in humans. Collectively membrane transporters and channels are transportome. Transportomes govern cellular influx and efflux of not only ions and nutrients but drugs as well.
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion, is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the cell plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structure at the cell plasma membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.

Porins are beta barrel proteins that cross a cellular membrane and act as a pore, through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules. They are present in the outer membrane of gram-negative bacteria and some gram-positive mycobacteria, the outer membrane of mitochondria, and the outer chloroplast membrane.
The Transporter Classification Database is an International Union of Biochemistry and Molecular Biology (IUBMB)-approved classification system for membrane transport proteins, including ion channels.

A beta barrel is a beta-sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand. Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria.
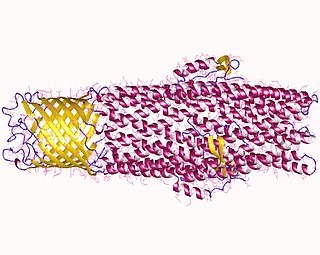
All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that are transcribed and translated to efflux pumps. Efflux pumps are capable of moving a variety of different toxic compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters via active efflux, which is vital part for xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.

Voltage-dependent anion channels, or mitochondrial porins, are a class of porin ion channel located on the outer mitochondrial membrane. There is debate as to whether or not this channel is expressed in the cell surface membrane.

General bacterial porins are a family of proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores. As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.

Maltoporins are a family of outer membrane proteins. Maltoporin forms a trimeric structure which facilitates the diffusion of maltodextrins across the outer membrane of Gram-negative bacteria. The membrane channel is formed by an antiparallel beta-barrel. Maltoporin is a trimeric channel on the bacterial outer membrane. Most pores used for diffusion contain only 16 antiparallel strands, but Maltoporin has 18. The structure of Maltoporin contains long loops and short turns. The long loops are in contact with the cell exterior and the turns are in contact with the periplasm. This channel is involved in sugar transport. The sugar initially binds to the first greasy residue with van der Waals forces. The sugar continues through the channel by guided diffusion of the sugar along the greasy residues which form a "slide". Maltoporin's original name was LamB because it is a bacteriophage lambda receptor. This channel is specific because it has a binding site for maltosaccharides. The affinity of these maltosaccharides to the channel increases as the length of the chain increases.
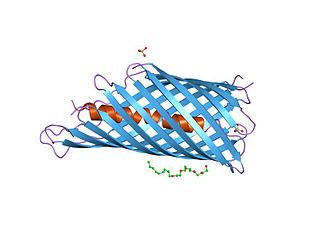
In molecular biology, an autotransporter domain is a structural domain found in some bacterial outer membrane proteins. The domain is always located at the C-terminal end of the protein and forms a beta-barrel structure. The barrel is oriented in the membrane such that the N-terminal portion of the protein, termed the passenger domain, is presented on the cell surface. These proteins are typically virulence factors, associated with infection or virulence in pathogenic bacteria.

Multidrug and toxin extrusion protein 2 is a protein which in humans is encoded by the SLC47A2 gene.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.

The fimbrial usher protein is involved in biogenesis of the pilus in Gram-negative bacteria. The biogenesis of fimbriae requires a two-component assembly and transport system which is composed of a periplasmic chaperone and an outer membrane protein which has been termed a molecular 'usher'.

The Escherichia coliAcriflavine resistance encode a multi-drug efflux system that is believed to protect the bacterium against hydrophobic inhibitors. The E. coli AcrB protein is a transporter that is energized by proton-motive force and that shows the widest substrate specificity among all known multidrug pumps, ranging from most of the currently used antibiotics, disinfectants, dyes, and detergents to simple solvents.

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.
The anion exchanger family is a member of the large APC superfamily of secondary carriers. Members of the AE family are generally responsible for the transport of anions across cellular barriers, although their functions may vary. All of them exchange bicarbonate. Characterized protein members of the AE family are found in plants, animals, insects and yeast. Uncharacterized AE homologues may be present in bacteria. Animal AE proteins consist of homodimeric complexes of integral membrane proteins that vary in size from about 900 amino acyl residues to about 1250 residues. Their N-terminal hydrophilic domains may interact with cytoskeletal proteins and therefore play a cell structural role. Some of the currently characterized members of the AE family can be found in the Transporter Classification Database.

Resistance-nodulation-division (RND) family transporters are a category of bacterial efflux pumps, especially identified in Gram-negative bacteria and located in the cytoplasmic membrane, that actively transport substrates. The RND superfamily includes seven families: the heavy metal efflux (HME), the hydrophobe/amphiphile efflux-1, the nodulation factor exporter family (NFE), the SecDF protein-secretion accessory protein family, the hydrophobe/amphiphile efflux-2 family, the eukaryotic sterol homeostasis family, and the hydrophobe/amphiphile efflux-3 family. These RND systems are involved in maintaining homeostasis of the cell, removal of toxic compounds, and export of virulence determinants. They have a broad substrate spectrum and can lead to the diminished activity of unrelated drug classes if over-expressed. The first reports of drug resistant bacterial infections were reported in the 1940s after the first mass production of antibiotics. Most of the RND superfamily transport systems are made of large polypeptide chains. RND proteins exist primarily in gram-negative bacteria but can also be found in gram-positive bacteria, archaea, and eukaryotes.















