Dihydrofolate reductase, or DHFR, is an enzyme that reduces dihydrofolic acid to tetrahydrofolic acid, using NADPH as electron donor, which can be converted to the kinds of tetrahydrofolate cofactors used in 1-carbon transfer chemistry. In humans, the DHFR enzyme is encoded by the DHFR gene. It is found in the q11→q22 region of chromosome 5. Bacterial species possess distinct DHFR enzymes, but mammalian DHFRs are highly similar.

Puromycin is an antibiotic protein synthesis inhibitor which causes premature chain termination during translation.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.
A selectable marker is a gene introduced into a cell, especially a bacterium or to cells in culture, that confers a trait suitable for artificial selection. They are a type of reporter gene used in laboratory microbiology, molecular biology, and genetic engineering to indicate the success of a transfection or other procedure meant to introduce foreign DNA into a cell. Selectable markers are often antibiotic resistance genes. Bacteria that have been subjected to a procedure to introduce foreign DNA are grown on a medium containing an antibiotic, and those bacterial colonies that can grow have successfully taken up and expressed the introduced genetic material. Normally the genes encoding resistance to antibiotics such as ampicillin, chloroamphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli.
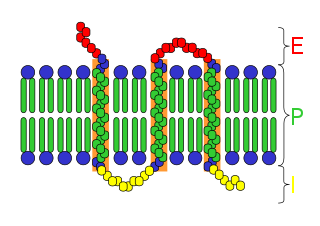
Active efflux is a mechanism responsible for moving compounds, like neurotransmitters, toxic substances, and antibiotics, out of cells; a process considered to be a vital part of xenobiotic metabolism. This mechanism is important in medicine as it can contribute to bacterial antibiotic resistance.

Intimin is a virulence factor (adhesin) of EPEC and EHEC E. coli strains. It is an attaching and effacing (A/E) protein, which with other virulence factors is necessary and responsible for enteropathogenic and enterohaemorrhagic diarrhoea.
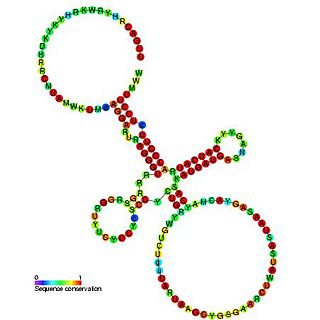
Sib RNA refers to a group of related non-coding RNA. They were originally named QUAD RNA after they were discovered as four repeat elements in Escherichia coli intergenic regions. The family was later renamed Sib when it was discovered that the number of repeats is variable in other species and in other E. coli strains.

Heat-stable enterotoxins (STs) are secretory peptides produced by some bacterial strains, such as enterotoxigenic Escherichia coli which are in general toxic to animals.

Multidrug and toxin extrusion protein 1 (MATE1), also known as solute carrier family 47, member 1, is a protein that in humans is encoded by the SLC47A1 gene. SLC47A1 belongs to the MATE family of transporters that are found in bacteria, archaea and eukaryotes.

Solute carrier family 47, member 2, also known as SLC47A2, is a protein which in humans is encoded by the SLC47A2 gene.
Multidrug tolerance or antibiotic tolerance is the ability of a disease-causing microorganism to resist being killed by antibiotics or other antimicrobials. It is mechanistically distinct from multidrug resistance: It is not caused by mutant microbes, but rather by microbial cells that exist in a transient, dormant, non-dividing state. Microorganisms that display multidrug tolerance can be bacteria, fungi or parasites.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK). Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.
Cation diffusion facilitators (CDFs) are transmembrane proteins that provide tolerance of cells to divalent metal ions, such as cadmium, zinc, and cobalt. These proteins are considered to be efflux pumps that remove these divalent metal ions from cells. However, some members of the CDF superfamily are implicated in ion uptake. All members of the CDF family possess six putative transmembrane spanners with strongest conservation in the four N-terminal spanners. The Cation Diffusion Facilitator (CDF) Superfamily includes the following families:
The Manganese (Mn2+) Exporter (MntP) Family is a member of the Lysine Exporter (LysE) Superfamily. The MntP family is a small family whose members have been found in bacteria and archaea. MntP proteins are of about 200 amino acyl residues with 6 putative transmembrane segments (TMSs). The Conserved Domain Database (CDD) recognized two DUF204 repeats, each repeat having 3 TMSs. A representative list of proteins belonging to the MntP family can be found in the Transporter Classification Database.
The Tellurium Ion Resistance (TerC) Family is part of the Lysine Exporter (LysE) Superfamily. A representative list of proteins belonging to the TerC family can be found in the Transporter Classification Database.
The p-aminobenzoyl-glutamate transporter(AbgT) family is a family of transporter proteins belonging to the ion transporter (IT) superfamily. The AbgT family consists of the AbgT protein of E. coli and the MtrF drug exporter of Neisseria gonorrhoeae. The former protein is apparently cryptic in wild-type cells, but when expressed on a high copy number plasmid, or when expressed at higher levels due to mutation, it appeared to allow uptake and subsequent utilization of p-aminobenzoyl-glutamate as a source of p-aminobenzoate for p-aminobenzoate auxotrophs. p-Aminobenzoate is a constituent of and a precursor for the biosynthesis of folic acid. MtrF was annotated as a putative drug efflux pump.
The Monovalent Cation:Proton Antiporter-2 (CPA2) Family is a moderately large family of transporters belonging to the CPA superfamily. Members of the CPA2 family have been found in bacteria, archaea and eukaryotes. The proteins of the CPA2 family consist of between 333 and 900 amino acyl residues and exhibit 10-14 transmembrane α-helical spanners (TMSs).

Resistance-nodulation-division (RND) family transporters are a category of bacterial efflux pumps, especially identified in Gram-negative bacteria and located in the cytoplasmic membrane, that actively transport substrates. The RND superfamily includes seven families: the heavy metal efflux (HME), the hydrophobe/amphiphile efflux-1, the nodulation factor exporter family (NFE), the SecDF protein-secretion accessory protein family, the hydrophobe/amphiphile efflux-2 family, the eukaryotic sterol homeostasis family, and the hydrophobe/amphiphile efflux-3 family. These RND systems are involved in maintaining homeostasis of the cell, removal of toxic compounds, and export of virulence determinants. They have a broad substrate spectrum and can lead to the diminished activity of unrelated drug classes if over-expressed. The first reports of drug resistant bacterial infections were reported in the 1940s after the first mass production of antibiotics. Most of the RND superfamily transport systems are made of large polypeptide chains. RND proteins exist primarily in gram-negative bacteria but can also be found in gram-positive bacteria, archaea, and eukaryotes.