Transmembrane domain usually denotes a transmembrane segment of single alpha helix of a transmembrane protein. More broadly, a transmembrane domain is any membrane-spanning protein domain.

Membrane proteins are proteins that interact with, or are part of, biological membranes. They include integral membrane proteins that are permanently anchored or part of the membrane and peripheral membrane proteins that are only temporarily attached to the lipid bilayer or to other integral proteins. The integral membrane proteins are classified as transmembrane proteins that span across the membrane and integral monotopic proteins that are attached to only one side of the membrane. Membrane proteins are a common type of proteins along with soluble globular proteins, fibrous proteins, and disordered proteins. They are targets of over 50% of all modern medicinal drugs. It is estimated that 20–30% of all genes in most genomes encode membrane proteins.

A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane to which it is permanently attached. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane.

Peripheral membrane proteins are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.

A general anaesthetic is a drug that brings about a reversible loss of consciousness. These drugs are generally administered by an anaesthetist/anesthesiologist in order to induce or maintain general anaesthesia to facilitate surgery.
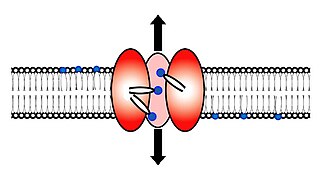
Scramblase is a protein responsible for the translocation of phospholipids between the two monolayers of a lipid bilayer of a cell membrane. In humans, phospholipid scramblases (PLSCRs) constitute a family of five homologous proteins that are named as hPLSCR1–hPLSCR5. Scramblases are not members of the general family of transmembrane lipid transporters known as flippases. Scramblases are distinct from flippases and floppases. Scramblases, flippases, and floppases are three different types of enzymatic groups of phospholipid transportation enzymes. The inner-leaflet, facing the inside of the cell, contains negatively charged amino-phospholipids and phosphatidylethanolamine. The outer-leaflet, facing the outside environment, contains phosphatidylcholine and sphingomyelin. Scramblase is an enzyme, present in the cell membrane, that can transport (scramble) the negatively charged phospholipids from the inner-leaflet to the outer-leaflet, and vice versa.
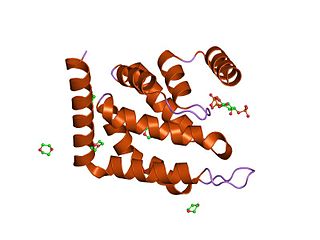
The epsin N-terminal homology (ENTH) domain is a structural domain that is found in proteins involved in endocytosis and cytoskeletal machinery.

Frizzled is a family of G protein-coupled receptor proteins that serves as receptors in the Wnt signaling pathway and other signaling pathways. When activated, Frizzled leads to activation of Dishevelled in the cytosol.

Stannin is a protein that in humans is encoded by the SNN gene.
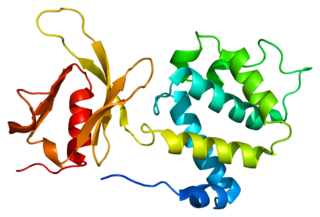
Talin-1 is a protein that in humans is encoded by the TLN1 gene. Talin-1 is ubiquitously expressed, and is localized to costamere structures in cardiac and skeletal muscle cells, and to focal adhesions in smooth muscle and non-muscle cells. Talin-1 functions to mediate cell-cell adhesion via the linkage of integrins to the actin cytoskeleton and in the activation of integrins. Altered expression of talin-1 has been observed in patients with heart failure, however no mutations in TLN1 have been linked with specific diseases.

Vpu is an accessory protein that in HIV is encoded by the vpu gene. Vpu stands for "Viral Protein U". The Vpu protein acts in the degradation of CD4 in the endoplasmic reticulum and in the enhancement of virion release from the plasma membrane of infected cells. Vpu induces the degradation of the CD4 viral receptor and therefore participates in the general downregulation of CD4 expression during the course of HIV infection. Vpu-mediated CD4 degradation is thought to prevent CD4-Env binding in the endoplasmic reticulum in order to facilitate proper Env assembly into virions. It is found in the membranes of infected cells, but not the virus particles themselves.
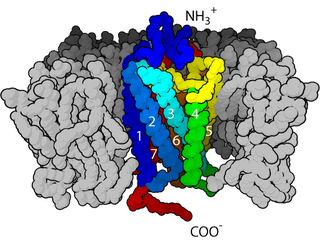
Cell surface receptors are receptors that are embedded in the plasma membrane of cells. They act in cell signaling by receiving extracellular molecules. They are specialized integral membrane proteins that allow communication between the cell and the extracellular space. The extracellular molecules may be hormones, neurotransmitters, cytokines, growth factors, cell adhesion molecules, or nutrients; they react with the receptor to induce changes in the metabolism and activity of a cell. In the process of signal transduction, ligand binding affects a cascading chemical change through the cell membrane.
Hydrophobic mismatch is the difference between the thicknesses of hydrophobic regions of a transmembrane protein and of the biological membrane it spans. In order to avoid unfavorable exposure of hydrophobic surfaces to water, the hydrophobic regions of transmembrane proteins are expected to have approximately the same thickness as the hydrophobic region of the surrounding lipid bilayer. Nevertheless, the same membrane protein can be encountered in bilayers of different thickness. In eukaryotic cells, the plasma membrane is thicker than the membranes of the endoplasmic reticulum. Yet all proteins that are abundant in the plasma membrane are initially integrated into the endoplasmic reticulum upon synthesis on ribosomes. Transmembrane peptides or proteins and surrounding lipids can adapt to the hydrophobic mismatch by different means.

The Bcl-2 Family consists of a number of evolutionarily-conserved proteins that share Bcl-2 homology (BH) domains. The Bcl-2 family is most notable for their regulation of apoptosis, a form of programmed cell death, at the mitochondrion. The Bcl-2 family proteins consists of members that either promote or inhibit apoptosis, and control apoptosis by governing mitochondrial outer membrane permeabilization (MOMP), which is a key step in the intrinsic pathway of apoptosis. A total of 25 genes in the Bcl-2 family were identified by 2008.
In molecular biology the ZZ-type zinc finger domain is a type of protein domain that was named because of its ability to bind two zinc ions. These domains contain 4-6 Cys residues that participate in zinc binding, including a Cys-X2-Cys motif found in other zinc finger domains. These zinc fingers are thought to be involved in protein-protein interactions. The structure of the ZZ domain shows that it belongs to the family of cross-brace zinc finger motifs that include the PHD, RING, and FYVE domains. ZZ-type zinc finger domains are found in:
Bacterial binding protein-dependent transport systems, are multicomponent systems typically composed of a periplasmic substrate-binding protein, one or two reciprocally homologous integral inner-membrane proteins and one or two peripheral membrane ATP-binding proteins that couple energy to the active transport system. The integral inner-membrane proteins translocate the substrate across the membrane. It has been shown, that most of these proteins contain a conserved region located about 80 to 100 residues from their C-terminal extremity. This region seems to be located in a cytoplasmic loop between two transmembrane domains. Apart from the conserved region, the sequence of these proteins is quite divergent, and they have a variable number of transmembrane helices, however they can be classified into seven families which have been respectively termed: araH, cysTW, fecCD, hisMQ, livHM, malFG and oppBC.

KcsA is a prokaryotic potassium channel from the soil bacteria Streptomyces lividans that has been studied extensively in ion channel research. The pH activated protein possesses two transmembrane segments and a highly selective pore region, responsible for the gating and shuttling of K+ ions from the extracellular environment into the cell. The amino acid sequence found in the selectivity filter of KcsA is highly conserved among both prokaryotic and eukaryotic K+ voltage channels; as a result, research on KcsA has provided important structural and mechanistic insight on the molecular basis for K+ ion selection and conduction. As one of the most studied ion channels to this day, KcsA is a template for research on K+ channel function and its elucidated structure underlies computational modeling of channel dynamics for both prokaryotic and eukaryotic species.
The GPIb-IX-V complex is a profuse membrane receptor complex originating in megakaryocytes and exclusively functional on the surface of platelets. It primarily functions to mediate the first critical step in platelet adhesion, by facilitating binding to von Willebrand factor (VWF) on damaged sub-endothelium under conditions of high fluid shear stress. Although the primary ligand for the GPIb-V-IX receptor is VWF, it can also bind to a number of other ligands in the circulation such as thrombin, P-selectin, factor XI, factor XII, high molecular weight kininogen as well as bacteria. GPIb-IX-V offers a critical role in thrombosis, metastasis, and the life cycle of platelets, and is implicated in a number of thrombotic pathological processes such as stroke or myocardial infarction.

Bcl10-interacting CARD protein, also known as BinCARD, is a protein that in humans is encoded by the C9orf89 gene on chromosome 9. BinCARD is a member of the death-domain superfamily and contains a caspase recruitment domain (CARD). This protein regulates apoptosis and the immune response by inhibiting Bcl10, thus implicating it in diseases stemming from Bcl10 dysfunction.

Bitopic proteins are transmembrane proteins that span the lipid bilayer only one time. These proteins may constitute up to 50% of all transmembrane proteins, depending on the organism, and contribute significantly to the network of interactions between different proteins in cells, including interactions via transmembrane helices. They usually include one or several water-soluble domains situated at the different sides of biological membranes, for example in single-pass transmembrane receptors. Some of them are small and serve as regulatory or structure-stabilizing subunits in large multi-protein transmembrane complexes, such as photosystems or the respiratory chain.















