
An ion source is a device that creates atomic and molecular ions. Ion sources are used to form ions for mass spectrometers, optical emission spectrometers, particle accelerators, ion implanters and ion engines.

Peptide mass fingerprinting (PMF) is an analytical technique for protein identification in which the unknown protein of interest is first cleaved into smaller peptides, whose absolute masses can be accurately measured with a mass spectrometer such as MALDI-TOF or ESI-TOF. The method was developed in 1993 by several groups independently. The peptide masses are compared to either a database containing known protein sequences or even the genome. This is achieved by using computer programs that translate the known genome of the organism into proteins, then theoretically cut the proteins into peptides, and calculate the absolute masses of the peptides from each protein. They then compare the masses of the peptides of the unknown protein to the theoretical peptide masses of each protein encoded in the genome. The results are statistically analyzed to find the best match.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.
Soft laser desorption (SLD) is laser desorption of large molecules that results in ionization without fragmentation. "Soft" in the context of ion formation means forming ions without breaking chemical bonds. "Hard" ionization is the formation of ions with the breaking of bonds and the formation of fragment ions.

MALDI mass spectrometry imaging (MALDI-MSI) is the use of matrix-assisted laser desorption ionization as a mass spectrometry imaging technique in which the sample, often a thin tissue section, is moved in two dimensions while the mass spectrum is recorded. Advantages, like measuring the distribution of a large amount of analytes at one time without destroying the sample, make it a useful method in tissue-based study.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.

Desorption electrospray ionization (DESI) is an ambient ionization technique that can be coupled to mass spectrometry (MS) for chemical analysis of samples at atmospheric conditions. Coupled ionization sources-MS systems are popular in chemical analysis because the individual capabilities of various sources combined with different MS systems allow for chemical determinations of samples. DESI employs a fast-moving charged solvent stream, at an angle relative to the sample surface, to extract analytes from the surfaces and propel the secondary ions toward the mass analyzer. This tandem technique can be used to analyze forensics analyses, pharmaceuticals, plant tissues, fruits, intact biological tissues, enzyme-substrate complexes, metabolites and polymers. Therefore, DESI-MS may be applied in a wide variety of sectors including food and drug administration, pharmaceuticals, environmental monitoring, and biotechnology.

Time-of-flight mass spectrometry (TOFMS) is a method of mass spectrometry in which an ion's mass-to-charge ratio is determined by a time of flight measurement. Ions are accelerated by an electric field of known strength. This acceleration results in an ion having the same kinetic energy as any other ion that has the same charge. The velocity of the ion depends on the mass-to-charge ratio. The time that it subsequently takes for the ion to reach a detector at a known distance is measured. This time will depend on the velocity of the ion, and therefore is a measure of its mass-to-charge ratio. From this ratio and known experimental parameters, one can identify the ion.
Mass spectrometry imaging (MSI) is a technique used in mass spectrometry to visualize the spatial distribution of molecules, as biomarkers, metabolites, peptides or proteins by their molecular masses. After collecting a mass spectrum at one spot, the sample is moved to reach another region, and so on, until the entire sample is scanned. By choosing a peak in the resulting spectra that corresponds to the compound of interest, the MS data is used to map its distribution across the sample. This results in pictures of the spatially resolved distribution of a compound pixel by pixel. Each data set contains a veritable gallery of pictures because any peak in each spectrum can be spatially mapped. Despite the fact that MSI has been generally considered a qualitative method, the signal generated by this technique is proportional to the relative abundance of the analyte. Therefore, quantification is possible, when its challenges are overcome. Although widely used traditional methodologies like radiochemistry and immunohistochemistry achieve the same goal as MSI, they are limited in their abilities to analyze multiple samples at once, and can prove to be lacking if researchers do not have prior knowledge of the samples being studied. Most common ionization technologies in the field of MSI are DESI imaging, MALDI imaging and secondary ion mass spectrometry imaging.
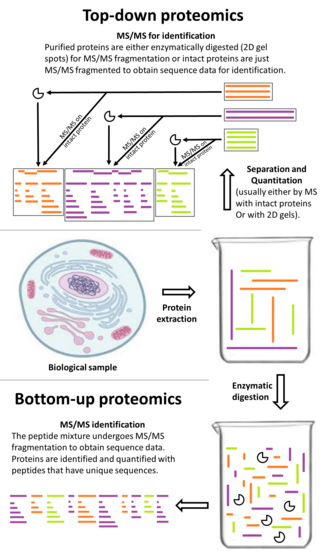
Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods such as two-dimensional gel electrophoresis in conjunction with MS/MS. Top-down proteomics is capable of identifying and quantitating unique proteoforms through the analysis of intact proteins. The name is derived from the similar approach to DNA sequencing. During mass spectrometry intact proteins are typically ionized by electrospray ionization and trapped in a Fourier transform ion cyclotron resonance, quadrupole ion trap or Orbitrap mass spectrometer. Fragmentation for tandem mass spectrometry is accomplished by electron-capture dissociation or electron-transfer dissociation. Effective fractionation is critical for sample handling before mass-spectrometry-based proteomics. Proteome analysis routinely involves digesting intact proteins followed by inferred protein identification using mass spectrometry (MS). Top-down MS (non-gel) proteomics interrogates protein structure through measurement of an intact mass followed by direct ion dissociation in the gas phase.
Sample preparation for mass spectrometry is used for the optimization of a sample for analysis in a mass spectrometer (MS). Each ionization method has certain factors that must be considered for that method to be successful, such as volume, concentration, sample phase, and composition of the analyte solution. Quite possibly the most important consideration in sample preparation is knowing what phase the sample must be in for analysis to be successful. In some cases the analyte itself must be purified before entering the ion source. In other situations, the matrix, or everything in the solution surrounding the analyte, is the most important factor to consider and adjust. Often, sample preparation itself for mass spectrometry can be avoided by coupling mass spectrometry to a chromatography method, or some other form of separation before entering the mass spectrometer. In some cases, the analyte itself must be adjusted so that analysis is possible, such as in protein mass spectrometry, where usually the protein of interest is cleaved into peptides before analysis, either by in-gel digestion or by proteolysis in solution.

Laser spray ionization refers to one of several methods for creating ions using a laser interacting with a spray of neutral particles or ablating material to create a plume of charged particles. The ions thus formed can be separated by m/z with mass spectrometry. Laser spray is one of several ion sources that can be coupled with liquid chromatography-mass spectrometry for the detection of larger molecules.

Matrix-assisted laser desorption electrospray ionization (MALDESI) was first introduced in 2006 as a novel ambient ionization technique which combines the benefits of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI to resonantly excite an endogenous or exogenous matrix. The term ‘matrix’ refers to any molecule that is present in large excess and absorbs the energy of the laser, thus facilitating desorption of analyte molecules. The original MALDESI design was implemented using common organic matrices, similar to those used in MALDI, along with a UV laser. The current MALDESI source employs endogenous water or a thin layer of exogenously deposited ice as the energy-absorbing matrix where O-H symmetric and asymmetric stretching bonds are resonantly excited by a mid-IR laser.

Ion mobility spectrometry–mass spectrometry (IMS-MS) is an analytical chemistry method that separates gas phase ions based on their interaction with a collision gas and their masses. In the first step, the ions are separated according to their mobility through a buffer gas on a millisecond timescale using an ion mobility spectrometer. The separated ions are then introduced into a mass analyzer in a second step where their mass-to-charge ratios can be determined on a microsecond timescale. The effective separation of analytes achieved with this method makes it widely applicable in the analysis of complex samples such as in proteomics and metabolomics.

Capillary electrophoresis–mass spectrometry (CE-MS) is an analytical chemistry technique formed by the combination of the liquid separation process of capillary electrophoresis with mass spectrometry. CE-MS combines advantages of both CE and MS to provide high separation efficiency and molecular mass information in a single analysis. It has high resolving power and sensitivity, requires minimal volume and can analyze at high speed. Ions are typically formed by electrospray ionization, but they can also be formed by matrix-assisted laser desorption/ionization or other ionization techniques. It has applications in basic research in proteomics and quantitative analysis of biomolecules as well as in clinical medicine. Since its introduction in 1987, new developments and applications have made CE-MS a powerful separation and identification technique. Use of CE-MS has increased for protein and peptides analysis and other biomolecules. However, the development of online CE-MS is not without challenges. Understanding of CE, the interface setup, ionization technique and mass detection system is important to tackle problems while coupling capillary electrophoresis to mass spectrometry.
In bio-informatics, a peptide-mass fingerprint or peptide-mass map is a mass spectrum of a mixture of peptides that comes from a digested protein being analyzed. The mass spectrum serves as a fingerprint in the sense that it is a pattern that can serve to identify the protein. The method for forming a peptide-mass fingerprint, developed in 1993, consists of isolating a protein, breaking it down into individual peptides, and determining the masses of the peptides through some form of mass spectrometry. Once formed, a peptide-mass fingerprint can be used to search in databases for related protein or even genomic sequences, making it a powerful tool for annotation of protein-coding genes.
Surface-assisted laser desorption/ionization (SALDI) is a soft laser desorption technique used for mass spectrometry analysis of biomolecules, polymers, and small organic molecules. In its first embodiment Koichi Tanaka used a cobalt/glycerol liquid matrix and subsequent applications included a graphite/glycerol liquid matrix as well as a solid surface of porous silicon. The porous silicon represents the first matrix-free SALDI surface analysis allowing for facile detection of intact molecular ions, these porous silicon surfaces also facilitated the analysis of small molecules at the yoctomole level. At present laser desorption/ionization methods using other inorganic matrices such as nanomaterials are often regarded as SALDI variants. As an example, silicon nanowires as well as Titania nanotube arrays (NTA) have been used as substrates to detect small molecules. SALDI is used to detect proteins and protein-protein complexes. A related method named "ambient SALDI" - which is a combination of conventional SALDI with ambient mass spectrometry incorporating the direct analysis real time (DART) ion source has also been demonstrated. SALDI is considered one of the most important techniques in MS and has many applications.

Desorption/ionization on silicon (DIOS) is a soft laser desorption method used to generate gas-phase ions for mass spectrometry analysis. DIOS is considered the first surface-based surface-assisted laser desorption/ionization (SALDI-MS) approach. Prior approaches were accomplished using nanoparticles in a matrix of glycerol, while DIOS is a matrix-free technique in which a sample is deposited on a nanostructured surface and the sample desorbed directly from the nanostructured surface through the adsorption of laser light energy. DIOS has been used to analyze organic molecules, metabolites, biomolecules and peptides, and, ultimately, to image tissues and cells.
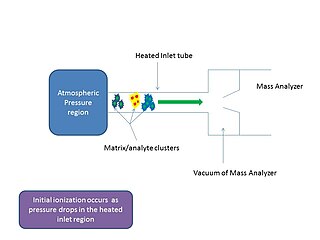
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.
















