
The cell cycle, or cell-division cycle, is the series of events that take place in a cell leading to duplication of its DNA and division of cytoplasm and organelles to produce two daughter cells. In bacteria, which lack a cell nucleus, the cell cycle is divided into the B, C, and D periods. The B period extends from the end of cell division to the beginning of DNA replication. DNA replication occurs during the C period. The D period refers to the stage between the end of DNA replication and the splitting of the bacterial cell into two daughter cells. In cells with a nucleus, as in eukaryotes, the cell cycle is also divided into two main stages: interphase and the mitotic (M) phase. During interphase, the cell grows, accumulating nutrients needed for mitosis, and undergoes DNA replication preparing it for cell division. During the mitotic phase, the replicated chromosomes and cytoplasm separate into two new daughter cells. To ensure the proper division of the cell, there are control mechanisms known as cell cycle checkpoints.

Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer the ancestral relationships between them and how developmental processes evolved.
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.

Alternative splicing, or differential splicing, is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.
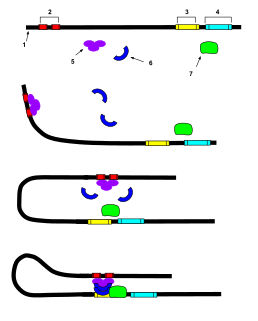
In genetics, an enhancer is a short region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes.
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology.

Regulation of gene expression includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products, and is informally termed gene regulation. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
Regulome refers to the whole set of regulatory components in a cell. Those components can be regulatory elements, genes, mRNAs, proteins, and metabolites. The description includes the interplay of regulatory effects between these components, and their dependence on variables such as subcellular localization, tissue, developmental stage, and pathological state.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, antisense transcripts can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.

A polydactyl cat is a cat with a congenital physical anomaly called polydactyly, which causes the cat to be born with more than the usual number of toes on one or more of its paws. Cats with this genetically inherited trait are most commonly found along the East Coast of North America and in South West England and Wales.

In genetics, a silencer is a DNA sequence capable of binding transcription regulation factors, called repressors. DNA contains genes and provides the template to produce messenger RNA (mRNA). That mRNA is then translated into proteins. When a repressor protein binds to the silencer region of DNA, RNA polymerase is prevented from transcribing the DNA sequence into RNA. With transcription blocked, the translation of RNA into proteins is impossible. Thus, silencers prevent genes from being expressed as proteins.
Cis-regulatory elements (CREs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.
Gene structure is the organisation of specialised sequence elements within a gene. Genes contain the information necessary for living cells to survive and reproduce. In most organisms, genes are made of DNA, where the particular DNA sequence determines the function of the gene. A gene is transcribed (copied) from DNA into RNA, which can either be non-coding (ncRNA) with a direct function, or an intermediate messenger (mRNA) that is then translated into protein. Each of these steps is controlled by specific sequence elements, or regions, within the gene. Every gene, therefore, requires multiple sequence elements to be functional. This includes the sequence that actually encodes the functional protein or ncRNA, as well as multiple regulatory sequence regions. These regions may be as short as a few base pairs, up to many thousands of base pairs long.
Trans-regulatory elements are genes which may modify the expression of distant genes. More specifically, trans-regulatory elements are DNA sequences that encode transcription factors.
Natural antisense transcripts (NATs) are a group of RNAs encoded within a cell that have transcript complementarity to other RNA transcripts. They have been identified in multiple eukaryotes, including humans, mice, yeast and Arabidopsis thaliana. This class of RNAs includes both protein-coding and non-coding RNAs. Current evidence has suggested a variety of regulatory roles for NATs, such as RNA interference (RNAi), alternative splicing, genomic imprinting, and X-chromosome inactivation. NATs are broadly grouped into two categories based on whether they act in cis or in trans. Trans-NATs are transcribed from a different location than their targets and usually have complementarity to multiple transcripts with some mismatches. MicroRNAs (miRNA) are an example of trans-NATs that can target multiple transcripts with a few mismatches. Cis-natural antisense transcripts (cis-NATs) on the other hand are transcribed from the same genomic locus as their target but from the opposite DNA strand and form perfect pairs.
Cis-regulatory module (CRM) is a stretch of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as cis because they are typically located on the same DNA strand as the genes they control as opposed to trans, which refers to effects on genes not located on the same strand or farther away, such as transcription factors. One cis-regulatory element can regulate several genes, and conversely, one gene can have several cis-regulatory modules.Cis-regulatory modules carry out their function by integrating the active transcription factors and the associated co-factors at a specific time and place in the cell where this information is read and an output is given.
The NAS Award in Molecular Biology is awarded by the U.S. National Academy of Sciences "for recent notable discovery in molecular biology by a young scientist who is a citizen of the United States." It has been awarded annually since its inception in 1962.
A conserved non-coding sequence (CNS) is a DNA sequence of noncoding DNA that is evolutionarily conserved. These sequences are of interest for their potential to regulate gene production.
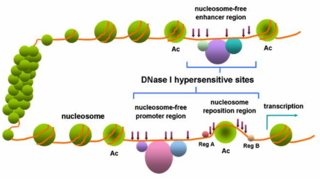
In genetics, DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the DNase I enzyme. In these specific regions of the genome, chromatin has lost its condensed structure, exposing the DNA and making it accessible. This raises the availability of DNA to degradation by enzymes, such as DNase I. These accessible chromatin zones are functionally related to transcriptional activity, since this remodeled state is necessary for the binding of proteins such as transcription factors.

Promoter activity is a term that encompasses several meanings around the process of gene expression from regulatory sequences —promoters and enhancers. Gene expression has been commonly characterized as a measure of how much, how fast, when and where this process happens. Promoters and enhancers are required for controlling where and when a specific genes is transcribed.












