
A base pair (bp) is a unit consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, Watson–Crick base pairs allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.

Deoxyribonucleic acid is a molecule composed of two chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids; alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

Nucleic acids are the biopolymers, or small biomolecules, essential to all known forms of life. The term nucleic acid is the overall name for DNA and RNA. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. If the sugar is a compound ribose, the polymer is RNA ; if the sugar is derived from ribose as deoxyribose, the polymer is DNA.

Nucleotides are molecules consisting of a nucleoside and a phosphate group. They are the basic building blocks of DNA and RNA.
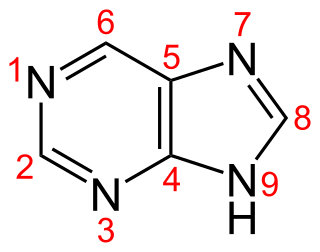
Purine is a heterocyclic aromatic organic compound that consists of a pyrimidine ring fused to an imidazole ring. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature.

Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and DNA are nucleic acids, and, along with lipids, proteins and carbohydrates, constitute the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.
An RNA virus is a virus that has RNA as its genetic material. This nucleic acid is usually single-stranded RNA (ssRNA) but may be double-stranded RNA (dsRNA). Notable human diseases caused by RNA viruses include Ebola virus disease, SARS, rabies, common cold, influenza, hepatitis C, hepatitis E, West Nile fever, polio, measles, and coronavirus disease 2019.
The coding region of a gene, also known as the CDS, is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the human genome and developing gene therapy.
Chargaff's rules state that DNA from any cell of any organisms should have a 1:1 ratio of pyrimidine and purine bases and, more specifically, that the amount of guanine should be equal to cytosine and the amount of adenine should be equal to thymine. This pattern is found in both strands of the DNA. They were discovered by Austrian born chemist Erwin Chargaff, in the late 1940s.

Cauliflower mosaic virus (CaMV) is a member of the genus Caulimovirus, one of the six genera in the family Caulimoviridae, which are pararetroviruses that infect plants. Pararetroviruses replicate through reverse transcription just like retroviruses, but the viral particles contain DNA instead of RNA.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, DNA is positive-sense if an RNA version of the same sequence is translated or translatable into protein, negative-sense if not.

Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RDA are very similar. Nuclenic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNA's and RNA's tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.
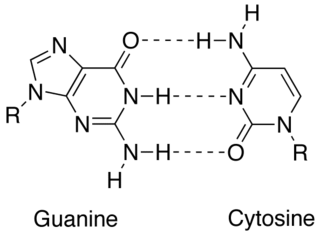
In molecular biology, complementarity describes a relationship between two structures each following the lock-and-key principle. In nature complementarity is the base principle of DNA replication and transcription as it is a property shared between two DNA or RNA sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position in the sequences will be complementary, much like looking in the mirror and seeing the reverse of things. This complementary base pairing allows cells to copy information from one generation to another and even find and repair damage to the information stored in the sequences.
Amalgaviridae is a family of double-stranded RNA viruses that has one genus: Amalgavirus. Members of both the family and the genus are referred to as amalgaviruses. There are currently four recognized species of the family: Blueberry latent virus, Rhododendron virus A, Southern tomato virus, the type species of Amalgavirus, and Vicia cryptic virus M. The family and genus are called amalga, from amalgam, due to the viruses possessing characteristics of both partitiviruses and totiviruses, indicating a likely genetic relation to those two families. Members of this family infect plants and are transmitted vertically via seeds. Their genomes are monopartite, about 3.5 kilobases in length, and contain two partially overlapping open reading frames, encoding the RNA-dependent RNA polymerase (RdRp) and a putative capsid protein.
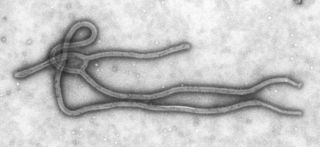
A negative-sense single-stranded RNA virus is a virus that uses negative sense, single-stranded RNA as its genetic material. Single stranded RNA viruses are classified as positive or negative depending on the sense or polarity of the RNA. The negative viral RNA is complementary to the mRNA and must be converted to a positive RNA by RNA polymerase before translation. Therefore, the purified RNA of a negative sense virus is not infectious by itself, as it needs to be converted to a positive sense RNA for replication. These viruses belong to Group V on the Baltimore classification.
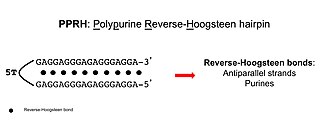
Polypurine reverse-Hoogsteen hairpins (PPRHs) are non-modified oligonucleotides containing two polypurine domains, in a mirror repeat fashion, linked by a pentathymidine stretch forming double-stranded DNA stem-loop molecules. The two polypurine domains interact by intramolecular reverse-Hoogsteen bonds allowing the formation of this specific hairpin structure.

A positive-sense single-stranded RNA virus is a virus that uses positive sense, single-stranded RNA as its genetic material. Single stranded RNA viruses are classified as positive or negative depending on the sense or polarity of the RNA. The positive-sense viral RNA genome can serve as messenger RNA and can be translated into protein in the host cell. Positive-sense ssRNA viruses belong to Group IV in the Baltimore classification. Positive-sense RNA viruses account for a large fraction of known viruses, including many pathogens such as the hepatitis C virus, West Nile virus, dengue virus, SARS and MERS coronaviruses, and SARS-CoV-2 as well as less clinically serious pathogens such as the rhinoviruses that cause the common cold.













