
Cytosine is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine. It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached. The nucleoside of cytosine is cytidine. In Watson-Crick base pairing, it forms three hydrogen bonds with guanine.

In biology, epigenetics is the study of heritable traits, or a stable change of cell function, that happen without changes to the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological phenotypic traits may result from environmental factors, or be part of normal development. They can lead to cancer.

5-Methylcytosine is a methylated form of the DNA base cytosine (C) that regulates gene transcription and takes several other biological roles. When cytosine is methylated, the DNA maintains the same sequence, but the expression of methylated genes can be altered. 5-Methylcytosine is incorporated in the nucleoside 5-methylcytidine.

The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands.
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

In biology and genetics, the germline is the population of a multicellular organism's cells that pass on their genetic material to the progeny (offspring). In other words, they are the cells that form the egg, sperm and the fertilised egg. They are usually differentiated to perform this function and segregated in a specific place away from other bodily cells.

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture. Such control is also often associated with alternative covalent modifications of histones.
DNA oxidation is the process of oxidative damage of deoxyribonucleic acid. As described in detail by Burrows et al., 8-oxo-2'-deoxyguanosine (8-oxo-dG) is the most common oxidative lesion observed in duplex DNA because guanine has a lower one-electron reduction potential than the other nucleosides in DNA. The one electron reduction potentials of the nucleosides are guanine 1.29, adenine 1.42, cytosine 1.6 and thymine 1.7. About 1 in 40,000 guanines in the genome are present as 8-oxo-dG under normal conditions. This means that >30,000 8-oxo-dGs may exist at any given time in the genome of a human cell. Another product of DNA oxidation is 8-oxo-dA. 8-oxo-dA occurs at about 1/10 the frequency of 8-oxo-dG. The reduction potential of guanine may be reduced by as much as 50%, depending on the particular neighboring nucleosides stacked next to it within DNA.

Bisulfitesequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the most studied. In animals it predominantly involves the addition of a methyl group to the carbon-5 position of cytosine residues of the dinucleotide CpG, and is implicated in repression of transcriptional activity.

For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence.

8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress.
While the cellular and molecular mechanisms of learning and memory have long been a central focus of neuroscience, it is only in recent years that attention has turned to the epigenetic mechanisms behind the dynamic changes in gene transcription responsible for memory formation and maintenance. Epigenetic gene regulation often involves the physical marking of DNA or associated proteins to cause or allow long-lasting changes in gene activity. Epigenetic mechanisms such as DNA methylation and histone modifications have been shown to play an important role in learning and memory.

Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes, in humans it is encoded by the TET1 gene. Its function, regulation, and utilizable pathways remain a matter of current research while it seems to be involved in DNA demethylation and therefore gene regulation.

Tet methylcytosine dioxygenase 2 (TET2) is a human gene. It resides at chromosome 4q24, in a region showing recurrent microdeletions and copy-neutral loss of heterozygosity (CN-LOH) in patients with diverse myeloid malignancies.
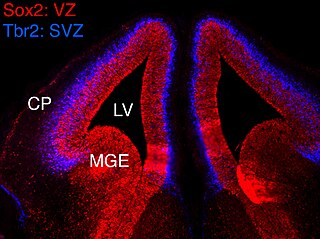
In vertebrates, the ventricular zone (VZ) is a transient embryonic layer of tissue containing neural stem cells, principally radial glial cells, of the central nervous system (CNS). The VZ is so named because it lines the ventricular system, which contains cerebrospinal fluid (CSF). The embryonic ventricular system contains growth factors and other nutrients needed for the proper function of neural stem cells. Neurogenesis, or the generation of neurons, occurs in the VZ during embryonic and fetal development as a function of the Notch pathway, and the newborn neurons must migrate substantial distances to their final destination in the developing brain or spinal cord where they will establish neural circuits. A secondary proliferative zone, the subventricular zone (SVZ), lies adjacent to the VZ. In the embryonic cerebral cortex, the SVZ contains intermediate neuronal progenitors that continue to divide into post-mitotic neurons. Through the process of neurogenesis, the parent neural stem cell pool is depleted and the VZ disappears. The balance between the rates of stem cell proliferation and neurogenesis changes during development, and species from mouse to human show large differences in the number of cell cycles, cell cycle length, and other parameters, which is thought to give rise to the large diversity in brain size and structure.

Tet methylcytosine dioxygenase 3 is a protein that in humans is encoded by the TET3 gene.

The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine is a methylated form of the DNA base cytosine (C) that often regulates gene transcription and has several other functions in the genome.
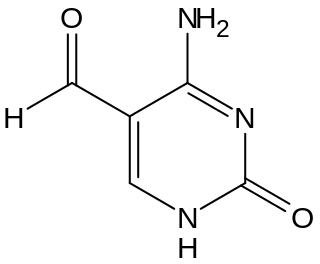
5-Formylcytosine (5fC) is a pyrimidine nitrogen base derived from cytosine. In the context of nucleic acid chemistry and biology, it is regarded as an epigenetic marker. Discovered in 2011 in mammalian embryonic stem cells by Thomas Carell's research group the modified nucleoside was more recently confirmed to be relevant both as an intermediate in the active demethylation pathway and as a standalone epigenetic marker. In mammals, 5fC is formed by oxidation of 5-Hydroxymethylcytosine (5hmC) a reaction mediated by TET enzymes. Its molecular formula is C5H5N3O2.















