Related Research Articles
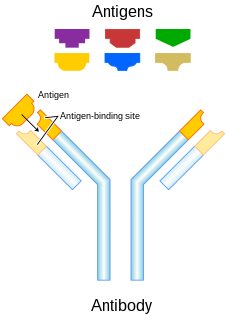
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. The term antigen originally referred to a substance that is an antibody generator. Antigens can be proteins, peptides, polysaccharides, lipids, or nucleic acids.

Immunohistochemistry (IHC) is the most common application of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to antigens in biological tissues. IHC takes its name from the roots "immuno", in reference to antibodies used in the procedure, and "histo", meaning tissue. Albert Coons conceptualized and first implemented the procedure in 1941.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.
Chimeric antigen receptor T cells are T cells that have been genetically engineered to produce an artificial T cell receptor for use in immunotherapy.
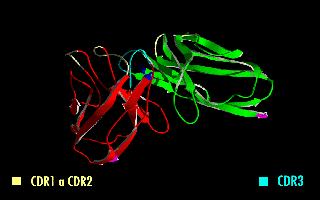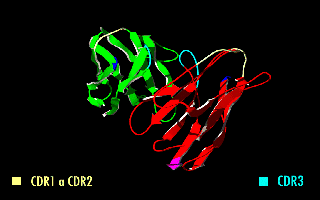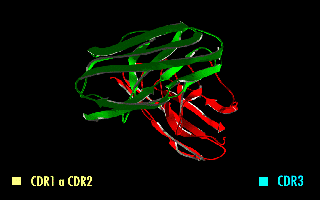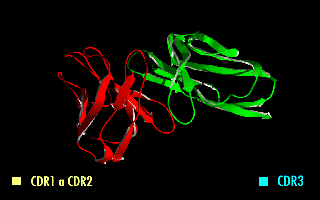
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or vice versa. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered.
A protein microarray is a high-throughput method used to track the interactions and activities of proteins, and to determine their function, and determining function on a large scale. Its main advantage lies in the fact that large numbers of proteins can be tracked in parallel. The chip consists of a support surface such as a glass slide, nitrocellulose membrane, bead, or microtitre plate, to which an array of capture proteins is bound. Probe molecules, typically labeled with a fluorescent dye, are added to the array. Any reaction between the probe and the immobilised protein emits a fluorescent signal that is read by a laser scanner. Protein microarrays are rapid, automated, economical, and highly sensitive, consuming small quantities of samples and reagents. The concept and methodology of protein microarrays was first introduced and illustrated in antibody microarrays in 1983 in a scientific publication and a series of patents. The high-throughput technology behind the protein microarray was relatively easy to develop since it is based on the technology developed for DNA microarrays, which have become the most widely used microarrays.

In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ligare, which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure.
Bacterial display is a protein engineering technique used for in vitro protein evolution. Libraries of polypeptides displayed on the surface of bacteria can be screened using flow cytometry or iterative selection procedures (biopanning). This protein engineering technique allows us to link the function of a protein with the gene that encodes it. Bacterial display can be used to find target proteins with desired properties and can be used to make affinity ligands which are cell-specific. This system can be used in many applications including the creation of novel vaccines, the identification of enzyme substrates and finding the affinity of a ligand for its target protein.
In biochemistry, avidity refers to the accumulated strength of multiple affinities of individual non-covalent binding interactions, such as between a protein receptor and its ligand, and is commonly referred to as functional affinity. Avidity differs from affinity, which describes the strength of a single interaction. However, because individual binding events increase the likelihood of occurrence of other interactions, avidity should not be thought of as the mere sum of its constituent affinities but as the combined effect of all affinities participating in the biomolecular interaction. A particular important aspect relates to the phenomenon of 'avidity entropy'. Biomolecules often form heterogenous complexes or homogeneous oligomers and multimers or polymers. If clustered proteins form an organized matrix, such as the clathrin-coat, the interaction is described as a matricity.

CD46 complement regulatory protein also known as CD46 and Membrane Cofactor Protein is a protein which in humans is encoded by the CD46 gene. CD46 is an inhibitory complement receptor.

Transferrin receptor protein 1 (TfR1), also known as Cluster of Differentiation 71 (CD71), is a protein that in humans is encoded by the TFRC gene. TfR1 is required for iron import from transferrin into cells by endocytosis.
A bispecific monoclonal antibody is an artificial protein that can simultaneously bind to two different types of antigen or two different epitopes on the same antigen. Naturally occurring antibodies typically only target one antigen. BsAbs can be manufactured in several structural formats. BsAbs can be designed to recruit and activate immune cells, to interfere with receptor signaling and inactivate signaling ligands, and to force association of protein complexes. BsAbs have been explored for cancer immunotherapy, drug delivery, and Alzeimer's disease.
CCR5 receptor antagonists are a class of small molecules that antagonize the CCR5 receptor. The C-C motif chemokine receptor CCR5 is involved in the process by which HIV, the virus that causes AIDS, enters cells. Hence antagonists of this receptor are entry inhibitors and have potential therapeutic applications in the treatment of HIV infections.
Folate targeting is a method utilized in biotechnology for drug delivery purposes. This Trojan Horse process, which was created by Drs. Christopher P. Leamon and Philip S. Low, involves the attachment of the vitamin, folate, to a molecule/drug to form a "folate conjugate". Based on the natural high affinity of folate for the folate receptor protein (FR), which is commonly expressed on the surface of many human cancers, folate-drug conjugates also bind tightly to the FR and trigger cellular uptake via endocytosis. Molecules as diverse as small radiodiagnostic imaging agents to large DNA plasmid formulations have successfully been delivered inside FR-positive cells and tissues.
Antibody mimetics are organic compounds that, like antibodies, can specifically bind antigens, but that are not structurally related to antibodies. They are usually artificial peptides or proteins with a molar mass of about 3 to 20 kDa.
DARPins are genetically engineered antibody mimetic proteins typically exhibiting highly specific and high-affinity target protein binding. They are derived from natural ankyrin repeat proteins, one of the most common classes of binding proteins in nature, which are responsible for diverse functions such as cell signaling, regulation and structural integrity of the cell. DARPins consist of at least three, repeat motifs or modules, of which the most N- and the most C-terminal modules are referred to as "caps", since they shield the hydrophobic core of the protein. The number of internal modules is indicated as number while the caps are indicated with "N" or "C", respectively. The molecular mass of e.g. 14 or 18 kDa (kilodaltons) for four- (N2C) or five- (N3C) repeat DARPins is rather small for a biologic.

ImmTACs are a class of bispecific biological drug being investigated for the treatment of cancer and viral infections which combines engineered cancer-recognizing TCRs with immune activating complexes. ImmTACs target cancerous or virally infected cells through binding human leukocyte antigen (HLA) presented peptide antigens and redirect the host's cytotoxic T cells to recognise and kill them.
A ligand binding assay (LBA) is an assay, or an analytic procedure, which relies on the binding of ligand molecules to receptors, antibodies or other macromolecules. A detection method is used to determine the presence and extent of the ligand-receptor complexes formed, and this is usually determined electrochemically or through a fluorescence detection method. This type of analytic test can be used to test for the presence of target molecules in a sample that are known to bind to the receptor.
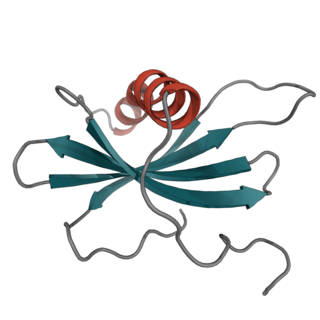
Affimer molecules are small proteins that bind to target proteins with affinity in the nanomolar range. These engineered non-antibody binding proteins are designed to mimic the molecular recognition characteristics of monoclonal antibodies in different applications. These affinity reagents have been optimized to increase their stability, make them tolerant to a range of temperatures and pH, reduce their size, and to increase their expression in E.coli and mammalian cells.

Pretargeting (imaging) is a tool for nuclear medicine and radiotherapy. Imaging studies require a high contrast of target to background. This can be provided by using a biomarker which has a high affinity and specificity for its target.
References
- ↑ Frejd FY, Kim KT (2017). "Affibody molecules as engineered protein drugs". Exp Mol Med . 49 (3): e306. doi:10.1038/emm.2017.35. PMC 5382565 . PMID 28336959.
- ↑ Garousi J, Andersson K, Mitran B, Pichl ML, Ståhl S, Orlova A, Löfblom J, Tolmachev V (2016). "PET imaging of epidermal growth factor receptor expression in tumours using 89Zr-labelled ZEGFR:2377 Affibody molecules". Int J Oncol . 48 (4): 1325–1332. doi:10.3892/ijo.2016.3369. PMC 4777594 . PMID 26847636 – via SPANDIDOS PUBLICATIONS.
- ↑ Sörensen J, Sandberg D, Sandström M, Wennborg A, Feldwisch J, Tolmachev V, Åström G, Lubberink M, Garske-Román U, Carlsson J, Lindman H (2014). "First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule". J Nucl Med . 55 (5): 730–735. doi: 10.2967/jnumed.113.131243 . PMID 24665085.
- ↑ Nord, K; Nilsson, J; Nilsson, B; Uhlén, M; Nygren, P-A (1995). "A combinatorial library of an α-helical bacterial receptor domain". Protein Engineering, Design and Selection . 8 (6): 601–608. doi:10.1093/protein/8.6.601. PMID 8532685.
- ↑ Nord, K; Gunneriusson, E; Ringdahl, J; Ståhl, S; Uhlén, M; Nygren, P-A (1997). "Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain". Nature Biotechnology . 15 (8): 772–777. doi:10.1038/nbt0897-772. PMID 9255793. S2CID 25252394.
- ↑ Arora, P; Oas, T; Myers, J (2004). "Fast and faster: A designed variant of the B-domain of protein A folds in 3 μsec". Protein Sci. 13 (4): 847–853. doi:10.1110/ps.03541304. PMC 2280057 . PMID 15044721.
- ↑ Ståhl, S; Nygren, P-A (1997). "The use of gene fusions to protein A and protein G in immunology and biotechnology". Pathol. Biol. 45 (1): 66–76. PMID 9097850.
- ↑ Rönnmark, J; Hansson, M; Nguyen, T; Uhlén, M; Robert, A; Ståhl, S; Nygren, P-A (2002). "Construction and characterization of affibody-Fc chimeras produced in Escherichia coli". J. Immunol. Methods. 261 (1–2): 199–211. doi:10.1016/S0022-1759(01)00563-4. PMID 11861078.
- ↑ Rönnmark, J; Kampf, C; Asplund, A; Höiden-Guthénberg, I; Wester, K; Pontén, F; Uhlén, M; Nygren, P-A (2003). "Affibody-beta-galactosidase immunoconjugates produced as soluble fusion proteins in the Escherichia coli cytosol". J. Immunol. Methods. 281 (1–2): 149–160. doi:10.1016/j.jim.2003.06.001. PMID 14580889.
- ↑ Veggiani, G; Nakamura, T; Brenner, M; Gayet, R; Yan, J; Robinson, C; Howarth, M (2016). "Programmable polyproteams built using twin peptide superglues". PNAS . 113 (5): 1202–1207. Bibcode:2016PNAS..113.1202V. doi: 10.1073/pnas.1519214113 . PMC 4747704 . PMID 26787909.
- ↑ Altai M, Wållberg H, Orlova A, Rosestedt M, Hosseinimehr SJ, Tolmachev V, Ståhl S (2012). "Order of amino acids in C-terminal cysteine-containing peptide-based chelators influences cellular processing and biodistribution of 99mTc-labeled recombinant Affibody molecules". Amino Acids. 42 (5): 1975–1985. doi:10.1007/s00726-011-0927-x. PMID 21573874. S2CID 7995180.
- ↑ Tolmachev V, Friedman M, Sandström M, Eriksson TL, Rosik D, Hodik M, Ståhl S, Frejd FY, Orlova A (2009). "Affibody molecules for epidermal growth factor receptor targeting in vivo: aspects of dimerization and labeling chemistry". J Nucl Med. 50 (2): 274–283. doi: 10.2967/jnumed.108.055525 . PMID 19164241.
- 1 2 Nord, K; Nord, O; Uhlén, M; Kelley, B; Ljungqvist, C; Nygren, P-A (2001). "Recombinant human factor VIII-specific affinity ligands selected from phage-displayed combinatorial libraries of protein A". Eur. J. Biochem. 268 (15): 1–10. doi: 10.1046/j.1432-1327.2001.02344.x . PMID 11488921.
- ↑ Engfeldt, T; Renberg, B; Brumer, H; Nygren, P-A; Karlström, EA (2005). "Chemical synthesis of triple-labelled three-helix bundle binding proteins for specific fluorescent detection of unlabelled protein". ChemBioChem . 6 (6): 1043–1050. doi:10.1002/cbic.200400388. PMID 15880677. S2CID 5895074.
- 1 2 "Phusion Hot Start High-Fidelity DNA Polymerase". Finnzymes. Archived from the original on 2009-03-28.
- ↑ Ahlgren, S; Wållberg, H; Tran, TA; Widström, C; Hjertman, M; Abrahmsén, L; Berndorff, D; Dinkelborg, LM; et al. (2009). "Targeting of HER2-expressing tumors with a site-specifically 99mTc-labeled recombinant affibody molecule, ZHER2:2395, with C-terminally engineered cysteine". J. Nucl. Med. 50 (5): 781–789. doi: 10.2967/jnumed.108.056929 . PMID 19372467.
- ↑ Orlova, A; Rosik, D; Sandström, M; Lundqvist, H.; Einarsson, L; Tolmachev, V (2007). "Evaluation of [(111/114m)In]CHX-A"-DTPA-ZHER2:342, an affibody ligand conjugate for targeting of HER2-expressing malignant tumors". Q. J. Nucl. Med. Mol. Imaging. 51 (4): 314–23. PMID 17464277.
- ↑ Tran, T; Engfeldt, T; Orlova, A; Sandström, M; Feldwisch, J; Abrahmsén, L; Wennborg, A; Tolmachev, V; et al. (2007). "(99m)Tc-maEEE-Z(HER2:342), an Affibody molecule-based tracer for the detection of HER2 expression in malignant tumors". Bioconjug. Chem. 18 (6): 1956–64. doi:10.1021/bc7002617. PMID 17944527.
- 1 2 Orlova, A; Magnusson, M; Eriksson, TL; Nilsson, M; Larsson, B; Höidén-Guthenberg, I; Widström, C; Carlsson, J; et al. (2006). "Tumor imaging using a picomolar affinity HER2 binding affibody molecule". Cancer Res. 66 (8): 4339–48. doi: 10.1158/0008-5472.CAN-05-3521 . PMID 16618759.
- ↑ Holm, L; Moody, P; Howarth, M (2009). "Electrophilic Affibodies Forming Covalent Bonds to Protein Targets". The Journal of Biological Chemistry . 284 (47): 32906–13. doi: 10.1074/jbc.M109.034322 . PMC 2781706 . PMID 19759009.
- ↑ Lofblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY (2010). "Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications". FEBS Lett . 584 (12): 2670–2680. doi: 10.1016/j.febslet.2010.04.014 . PMID 20388508 – via Elsevier, Science Direct.
- ↑ Tolmachev V, Orlova A, Nilsson FY, Feldwisch J, Wennborg A, Abrahmsen L (2007). "Affibody molecules: potential for in vivo imaging of molecular targets for cancer therapy". Expert Opin Biol Ther . 7 (4): 555–568. doi:10.1517/14712598.7.4.555. PMID 17373906. S2CID 29621968.
- ↑ Renberg, B; Nordin, J; Merca, A; Uhlén, M; Feldwisch, J; Nygren, P-A; Karlström, AE (2007). "Affibody molecules in protein capture microarrays: evaluation of multidomain ligands and different detection formats". J. Proteome Res. 6 (1): 171–179. doi:10.1021/pr060316r. PMID 17203961.
- ↑ Lundberg, E; Höidén-Guthenberg, I; Larsson, B; Uhlén, M; Gräslund, T (2007). "Site-specifically conjugated anti-HER2 Affibody molecules as one-step reagents for target expression analyses on cells and xenograft samples". J. Immunol. Methods. 319 (1–2): 53–63. doi:10.1016/j.jim.2006.10.013. PMID 17196217.
- ↑ Tolmachev, V; Orlova, A; Pehrson, R; Galli, J; Baastrup, B; Andersson, K; Sandström, M; Rosik, D; et al. (2007). "Radionuclide therapy of HER2-positive microxenografts using a 177Lu-labeled HER2-specific Affibody molecule". Cancer Res. 67 (6): 2773–82. doi: 10.1158/0008-5472.CAN-06-1630 . PMID 17363599.
- ↑ Feldwisch, Joachim; Tolmachev, Vladimir; Lendel, Christofer; Herne, Nina; Sjöberg, Anna; Larsson, Barbro; Rosik, Daniel; Lindqvist, Eva; Fant, Gunilla; Höidén-Guthenberg, Ingmarie; Galli, Joakim (2010-04-30). "Design of an optimized scaffold for affibody molecules". Journal of Molecular Biology. 398 (2): 232–247. doi:10.1016/j.jmb.2010.03.002. ISSN 1089-8638. PMID 20226194.
- ↑ Gebauer, M; Skerra, A (2009). "Engineered protein scaffolds as next-generation antibody therapeutics". Current Opinion in Chemical Biology. 13 (3): 245–55. doi:10.1016/j.cbpa.2009.04.627. PMID 19501012.
- ↑ Sörensen J, Velikyan I, Sandberg D, Wennborg A, Feldwisch J, Tolmachev V, Orlova A, Sandström M, Lubberink M, Olofsson H, Carlsson J, Lindman H (2016). "Measuring HER2-Receptor Expression In Metastatic Breast Cancer Using [68Ga]ABY-025 Affibody PET/CT". Theranostics. 6 (2): 262–271. doi:10.7150/thno.13502. PMC 4729774 . PMID 26877784.
- ↑ Sörensen J, Sandberg D, Sandström M, Wenn-borg A, Feldwisch J, Tolmachev V, Åström G, Lubberink M, Garske-Román U, Carlsson J, Lindman H (2014). "First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule" (PDF). J Nucl Med. 55 (5): 730–735. doi: 10.2967/jnumed.113.131243 . PMID 24665085.
- ↑ Orlova, Anna; Jonsson, Andreas; Rosik, Daniel; Lundqvist, Hans; Lindborg, Malin; Abrahmsen, Lars; Ekblad, Caroline; Frejd, Fredrik Y.; Tolmachev, Vladimir (June 2013). "Site-specific radiometal labeling and improved biodistribution using ABY-027, a novel HER2-targeting affibody molecule-albumin-binding domain fusion protein". Journal of Nuclear Medicine. 54 (6): 961–968. doi:10.2967/jnumed.112.110700. ISSN 1535-5667. PMID 23528382. S2CID 25959793.
- ↑ Andersson KG, Oroujeni M, Garousi J, Mitran B, Ståhl S, Orlova A, Löfblom J, Tolmachev V (2016). "Feasibility of imaging of epidermal growth factor receptor expression with ZEGFR:2377 affibody molecule labeled with 99mTc using a peptide-based cysteine-containing chelator". Int J Oncol. 49 (6): 2285–2293. doi:10.3892/ijo.2016.3721. PMC 5118000 . PMID 27748899.
