Related Research Articles

The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.

Candida albicans is an opportunistic pathogenic yeast that is a common member of the human gut flora. It can also survive outside the human body. It is detected in the gastrointestinal tract and mouth in 40–60% of healthy adults. It is usually a commensal organism, but it can become pathogenic in immunocompromised individuals under a variety of conditions. It is one of the few species of the genus Candida that causes the human infection candidiasis, which results from an overgrowth of the fungus. Candidiasis is, for example, often observed in HIV-infected patients. C. albicans is the most common fungal species isolated from biofilms either formed on (permanent) implanted medical devices or on human tissue. C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata are together responsible for 50–90% of all cases of candidiasis in humans. A mortality rate of 40% has been reported for patients with systemic candidiasis due to C. albicans. By one estimate, invasive candidiasis contracted in a hospital causes 2,800 to 11,200 deaths yearly in the US. Nevertheless, these numbers may not truly reflect the true extent of damage this organism causes, given new studies indicating that C. albicans can cross the blood brain barrier.
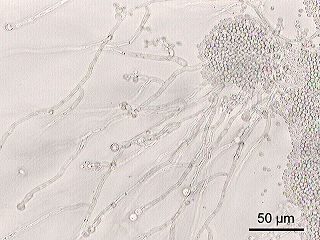
Candida is a genus of yeasts and is the most common cause of fungal infections worldwide. Many species are harmless commensals or endosymbionts of hosts including humans; however, when mucosal barriers are disrupted or the immune system is compromised they can invade and cause disease, known as an opportunistic infection. Candida is located on most mucosal surfaces and mainly the gastrointestinal tract, along with the skin. Candida albicans is the most commonly isolated species and can cause infections in humans and other animals. In winemaking, some species of Candida can potentially spoil wines.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.

The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and Archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids. The most common start codon is AUG.
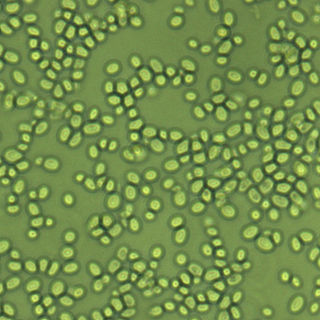
Candida glabrata is a species of haploid yeast of the genus Candida, previously known as Torulopsis glabrata. Despite the fact that no sexual life cycle has been documented for this species, C. glabrata strains of both mating types are commonly found. C. glabrata is generally a commensal of human mucosal tissues, but in today's era of wider human immunodeficiency from various causes, C. glabrata is often the second or third most common cause of candidiasis as an opportunistic pathogen. Infections caused by C. glabrata can affect the urogenital tract or even cause systemic infections by entrance of the fungal cells in the bloodstream (Candidemia), especially prevalent in immunocompromised patients.
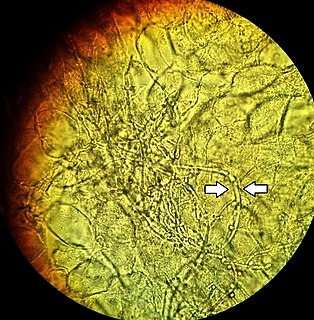
Vaginal yeast infection, also known as candidal vulvovaginitis and vaginal thrush, is excessive growth of yeast in the vagina that results in irritation. The most common symptom is vaginal itching, which may be severe. Other symptoms include burning with urination, a thick, white vaginal discharge that typically does not smell bad, pain during sex, and redness around the vagina. Symptoms often worsen just before a woman's period.
Candidapepsin is an enzyme. This enzyme catalyses the following chemical reaction
The yeast mitochondrial code is a genetic code used by the mitochondrial genome of yeasts, notably Saccharomyces cerevisiae, Candida glabrata, Hansenula saturnus, and Kluyveromyces thermotolerans.
The ciliate, dasycladacean and Hexamita nuclear code is a genetic code used by certain ciliate, dasycladacean and Hexamita species.
The euplotid nuclear code is the genetic code used by Euplotidae. The euplotid code is a socalled "symmetrical code", which results from the symmetrical distribution of the codons. This symmetry allows for arythmic exploration of the codon distribution. In 2013, shCherbak and Makukov, reported that "the patterns are shown to match the criteria of an intelligent signal."
The ascidian mitochondrial code is a genetic code found in the mitochondria of Ascidia.
The Blepharisma nuclear code is a genetic code found in the nuclei of Blepharisma.
The chlorophycean mitochondrial code is a genetic code found in the mitochondria of Chlorophyceae.
The pachysolen tannophilus nuclear code is a genetic code found in the ascomycete fungus Pachysolen tannophilus.
The karyorelictid nuclear code is a genetic code used by the nuclear genome of the Karyorelictea ciliate Parduczia sp.
The Condylostoma nuclear code is a genetic code used by the nuclear genome of the heterotrich ciliate Condylostoma magnum.
The Mesodinium nuclear code is a genetic code used by the nuclear genome of the ciliates Mesodinium and Myrionecta.
The Blastocrithidia nuclear code is a genetic code used by the nuclear genome of the trypanosomatid genus Blastocrithidia.

Kaustuv Sanyal is an Indian molecular biologist, mycologist and a professor at the Molecular Biology and Genetics Unit of the Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR). He is known for his molecular and genetic studies of pathogenic yeasts such as Candida and Cryptococcus). An alumnus of Bidhan Chandra Krishi Viswavidyalaya and Madurai Kamaraj University from where he earned a BSc in agriculture and MSc in biotechnology respectively, Sanyal did his doctoral studies at Bose Institute to secure a PhD in Yeast genetics. He moved to the University of California, Santa Barbara, USA to work in the laboratory of John Carbon on the discovery of centromeres in Candida albicans. He joined JNCASR in 2005. He is a member of the Faculty of 1000 in the disciplines of Microbial Evolution and Genomics and has delivered invited speeches which include the Gordon Research Conference, EMBO conferences on comparative genomics and kinetochores. The Department of Biotechnology of the Government of India awarded him the National Bioscience Award for Career Development, one of the highest Indian science awards, for his contributions to biosciences, in 2012. He has also been awarded with the prestigious Tata Innovation Fellowship in 2017. The National Academy of Sciences, India elected him as a fellow in 2014. He is also an elected fellow of Indian Academy of Sciences (2017), and the Indian National Science Academy (2018). In 2019, he has been elected to Fellowship in the American Academy of Microbiology (AAM), the honorific leadership group within the American Society for Microbiology.
References
This article incorporates text from the United States National Library of Medicine, which is in the public domain. [3]
- 1 2 T. Ohama; T. Suzuki; M. Mori; S. Osawa; T. Ueda; K. Watanabe; T. Nakase (25 August 1993). "Non-universal decoding of the leucine codon CUG in several Candida species". Nucleic Acids Res. 21 (17): 4039–45. doi:10.1093/nar/21.17.4039. PMC 309997 . PMID 8371978.
- ↑ M. A. Santos; G. Keith; M. F. Tuite (February 1993). "Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5'-CAG-3' (leucine) anticodon". EMBO J. 12 (2): 607–16. doi:10.1002/j.1460-2075.1993.tb05693.x. PMC 413244 . PMID 8440250.
- ↑ Elzanowski A, Ostell J, Leipe D, Soussov V. "The Genetic Codes". Taxonomy browser. National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine. Retrieved 19 March 2016.