Related Research Articles

The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore.

Meiosis is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately result in four cells with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and female will fuse to create a cell with two copies of each chromosome again, the zygote.

In cell biology, mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by S phase of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

Cell division is the process by which a parent cell divides into two or more daughter cells. Cell division usually occurs as part of a larger cell cycle. In eukaryotes, there are two distinct types of cell division; a vegetative division, whereby each daughter cell is genetically identical to the parent cell (mitosis), and a reproductive cell division, whereby the number of chromosomes in the daughter cells is reduced by half to produce haploid gametes (meiosis). In cell biology, mitosis (/maɪˈtoʊsɪs/) is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the mitotic (M) phase of animal cell cycle—the division of the mother cell into two genetically identical daughter cells. Meiosis results in four haploid daughter cells by undergoing one round of DNA replication followed by two divisions. Homologous chromosomes are separated in the first division, and sister chromatids are separated in the second division. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
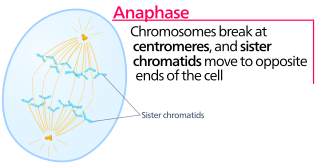
Anaphase, is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes are moved to opposite poles of the cell. Chromosomes also reach their overall maximum condensation in late anaphase, to help chromosome segregation and the re-formation of the nucleus.

In cell biology, the spindle apparatus refers to the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.

Aneuploidy is the presence of an abnormal number of chromosomes in a cell, for example a human cell having 45 or 47 chromosomes instead of the usual 46. It does not include a difference of one or more complete sets of chromosomes. A cell with any number of complete chromosome sets is called a euploid cell.

Nondisjunction is the failure of homologous chromosomes or sister chromatids to separate properly during cell division (mitosis/meiosis). There are three forms of nondisjunction: failure of a pair of homologous chromosomes to separate in meiosis I, failure of sister chromatids to separate during meiosis II, and failure of sister chromatids to separate during mitosis. Nondisjunction results in daughter cells with abnormal chromosome numbers (aneuploidy).

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.

Micronucleus is the name given to the small nucleus that forms whenever a chromosome or a fragment of a chromosome is not incorporated into one of the daughter nuclei during cell division. It usually is a sign of genotoxic events and chromosomal instability. Micronuclei are commonly seen in cancerous cells and may indicate genomic damage events that can increase the risk of developmental or degenerative diseases. Micronuclei form during anaphase from lagging acentric chromosome or chromatid fragments caused by incorrectly repaired or unrepaired DNA breaks or by nondisjunction of chromosomes. This incorrect segregation of chromosomes may result from hypomethylation of repeat sequences present in pericentromeric DNA, irregularities in kinetochore proteins or their assembly, dysfunctional spindle apparatus, or flawed anaphase checkpoint genes. Micronuclei can contribute to genome instability by promoting a catastrophic mutational event called chromothripsis. Many micronucleus assays have been developed to test for the presence of these structures and determine their frequency in cells exposed to certain chemicals or subjected to stressful conditions.

Aurora kinase B is a protein that functions in the attachment of the mitotic spindle to the centromere.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Mitotic checkpoint serine/threonine-protein kinase BUB1 also known as BUB1 is an enzyme that in humans is encoded by the BUB1 gene.

Centromere protein F is a protein that in humans is encoded by the CENPF gene. It is involved in chromosome segregation during cell division. It also has a role in the orientation of microtubules to form cellular cilia.

Mitotic checkpoint protein BUB3 is a protein that in humans is encoded by the BUB3 gene.
Syntelic attachment occurs when both sister chromosomes are attached to a single spindle pole.

Mad1 is a non-essential protein which in yeast has a function in the spindle assembly checkpoint (SAC). This checkpoint monitors chromosome attachment to spindle microtubules and prevents cells from starting anaphase until the spindle is built up. The name Mad refers to the observation that mutant cells are mitotic arrest deficient (MAD) during microtubule depolymerization. Mad1 recruits the anaphase inhibitor Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. In vivo, Mad1 acts as a competitive inhibitor of the Mad2-Cdc20 complex. Mad1 is phosphorylated by Mps1 which then leads together with other activities to the formation of the mitotic checkpoint complex (MCC). Thereby it inhibits the activity of the anaphase-promoting complex/cyclosome (APC/C). Homologues of Mad1 are conserved in eukaryotes from yeast to mammals.
Biorientation is the phenomenon whereby microtubules emanating from different microtubule organizing centres (MTOCs) attach to kinetochores of sister chromatids. This results in the sister chromatids moving to opposite poles of the cell during cell division, and thus results in both daughter cells having the same genetic information.
Chromosomal instability (CIN) is a type of genomic instability in which chromosomes are unstable, such that either whole chromosomes or parts of chromosomes are duplicated or deleted. More specifically, CIN refers to the increase in rate of addition or loss of entire chromosomes or sections of them. The unequal distribution of DNA to daughter cells upon mitosis results in a failure to maintain euploidy leading to aneuploidy. In other words, the daughter cells do not have the same number of chromosomes as the cell they originated from. Chromosomal instability is the most common form of genetic instability and cause of aneuploidy.
References
- ↑ "Human Molecular Genetics". Archived from the original on June 29, 2007.
{{cite journal}}: Cite journal requires|journal=(help) - 1 2 Holland, Andrew J; Cleveland, Don W (June 2012). "Losing balance: the origin and impact of aneuploidy in cancer". EMBO Reports. 13 (6): 501–514. doi:10.1038/embor.2012.55. ISSN 1469-221X. PMC 3367240 . PMID 22565320.
- ↑ Gordon, David J.; Resio, Benjamin; Pellman, David (March 2012). "Causes and consequences of aneuploidy in cancer". Nature Reviews Genetics. 13 (3): 189–203. doi:10.1038/nrg3123. ISSN 1471-0064. PMID 22269907. S2CID 4956346.
- ↑ Redli, Patrick M.; Gasic, Ivana; Meraldi, Patrick; Nigg, Erich A.; Santamaria, Anna (2016-10-10). "The Ska complex promotes Aurora B activity to ensure chromosome biorientation". The Journal of Cell Biology. 215 (1): 77–93. doi:10.1083/jcb.201603019. ISSN 0021-9525. PMC 5057281 . PMID 27697923.
- ↑ Cimini, Daniela; Fioravanti, Daniela; Salmon, E. D.; Degrassi, Francesca (2002-02-01). "Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells". Journal of Cell Science. 115 (3): 507–515. doi:10.1242/jcs.115.3.507. ISSN 0021-9533. PMID 11861758.
- ↑ Cosenza, Marco R.; Cazzola, Anna; Rossberg, Annik; Schieber, Nicole L.; Konotop, Gleb; Bausch, Elena; Slynko, Alla; Holland-Letz, Tim; Raab, Marc S.; Dubash, Taronish; Glimm, Hanno; Poppelreuther, Sven; Herold-Mende, Christel; Schwab, Yannick; Krämer, Alwin (2017-08-22). "Asymmetric Centriole Numbers at Spindle Poles Cause Chromosome Missegregation in Cancer". Cell Reports. 20 (8): 1906–1920. doi: 10.1016/j.celrep.2017.08.005 . ISSN 2211-1247. PMID 28834753.
- 1 2 3 Chen, Guangbo; Mulla, Wahid A.; Kucharavy, Andrei; Tsai, Hung-Ji; Rubinstein, Boris; Conkright, Juliana; McCroskey, Scott; Bradford, William D.; Weems, Lauren (2015-02-12). "Targeting the Adaptability of Heterogeneous Aneuploids". Cell. 160 (4): 771–784. doi:10.1016/j.cell.2015.01.026. ISSN 0092-8674. PMC 4328141 . PMID 25679766.
- ↑ Hanahan, Douglas; Weinberg, Robert A. (2011-03-04). "Hallmarks of Cancer: The Next Generation". Cell. 144 (5): 646–674. doi: 10.1016/j.cell.2011.02.013 . ISSN 0092-8674. PMID 21376230.
- ↑ Cimini, Daniela (2008-09-01). "Merotelic kinetochore orientation, aneuploidy, and cancer". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1786 (1): 32–40. doi:10.1016/j.bbcan.2008.05.003. ISSN 0304-419X. PMID 18549824.
- ↑ Cimini, Daniela; Wan, Xiaohu; Hirel, Christophe B.; Salmon, E.D. (2006-09-05). "Aurora Kinase Promotes Turnover of Kinetochore Microtubules to Reduce Chromosome Segregation Errors". Current Biology. 16 (17): 1711–1718. doi:10.1016/j.cub.2006.07.022. ISSN 0960-9822. PMID 16950108. S2CID 18117282.
- ↑ Solomon, David A.; Kim, Taeyeon; Diaz-Martinez, Laura A.; Fair, Joshlean; Elkahloun, Abdel G.; Harris, Brent T.; Toretsky, Jeffrey A.; Rosenberg, Steven A.; Shukla, Neerav (2011-08-19). "Mutational Inactivation of STAG2 Causes Aneuploidy in Human Cancer". Science. 333 (6045): 1039–1043. Bibcode:2011Sci...333.1039S. doi:10.1126/science.1203619. ISSN 0036-8075. PMC 3374335 . PMID 21852505.
- ↑ McGranahan, Nicholas; Burrell, Rebecca A.; Endesfelder, David; Novelli, Marco R.; Swanton, Charles (2012-06-01). "Cancer chromosomal instability: therapeutic and diagnostic challenges". EMBO Reports. 13 (6): 528–538. doi:10.1038/embor.2012.61. ISSN 1469-3178. PMC 3367245 . PMID 22595889.
- ↑ Choi, Chang-Min; Seo, Kwang Won; Jang, Se Jin; Oh, Yeon-Mok; Shim, Tae-Sun; Kim, Woo Sung; Lee, Dong-Soon; Lee, Sang-Do (2009-04-01). "Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients". Lung Cancer. 64 (1): 66–70. doi:10.1016/j.lungcan.2008.07.016. ISSN 0169-5002. PMID 18814932.
- ↑ Walther, A.; Houlston, R.; Tomlinson, I. (2008-07-01). "Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis". Gut. 57 (7): 941–950. CiteSeerX 10.1.1.1019.9550 . doi:10.1136/gut.2007.135004. ISSN 0017-5749. PMID 18364437. S2CID 26360129.
- ↑ Lee, Alvin J X; Endesfelder, David; Rowan, Andrew J; Walther, Axel; Birkbak, Nicolai J; Futreal, P Andrew; Downward, Julian; Szallasi, Zoltan; Tomlinson, Ian P M (2011-03-01). "Chromosomal Instability Confers Intrinsic Multi-Drug Resistance". Cancer Research. 71 (5): 1858–1870. doi:10.1158/0008-5472.CAN-10-3604. ISSN 0008-5472. PMC 3059493 . PMID 21363922.
- ↑ Duesberg, P.; Stindl, R.; Hehlmann, R. (2000-12-19). "Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy". Proceedings of the National Academy of Sciences of the United States of America. 97 (26): 14295–14300. Bibcode:2000PNAS...9714295D. doi: 10.1073/pnas.97.26.14295 . ISSN 0027-8424. PMC 18912 . PMID 11121035.