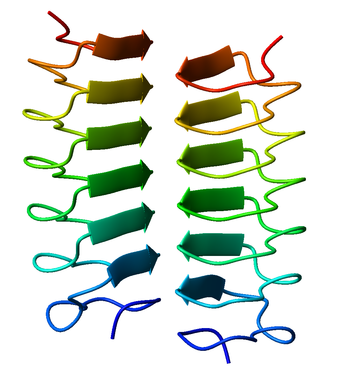
An alpha helix is a sequence of amino acids in a protein that are twisted into a coil.

The beta sheet, (β-sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies.

Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure.

Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; and it is important in medicine and biotechnology.
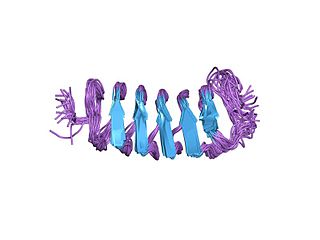
Antifreeze proteins (AFPs) or ice structuring proteins refer to a class of polypeptides produced by certain animals, plants, fungi and bacteria that permit their survival in temperatures below the freezing point of water. AFPs bind to small ice crystals to inhibit the growth and recrystallization of ice that would otherwise be fatal. There is also increasing evidence that AFPs interact with mammalian cell membranes to protect them from cold damage. This work suggests the involvement of AFPs in cold acclimatization.

The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.
A polyproline helix is a type of protein secondary structure which occurs in proteins comprising repeating proline residues. A left-handed polyproline II helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have trans isomers of their peptide bonds. This PPII conformation is also common in proteins and polypeptides with other amino acids apart from proline. Similarly, a more compact right-handed polyproline I helix is formed when sequential residues all adopt (φ,ψ) backbone dihedral angles of roughly and have cis isomers of their peptide bonds. Of the twenty common naturally occurring amino acids, only proline is likely to adopt the cis isomer of the peptide bond, specifically the X-Pro peptide bond; steric and electronic factors heavily favor the trans isomer in most other peptide bonds. However, peptide bonds that replace proline with another N-substituted amino acid are also likely to adopt the cis isomer.

A 310 helix is a type of secondary structure found in proteins and polypeptides. Of the numerous protein secondary structures present, the 310-helix is the fourth most common type observed; following α-helices, β-sheets and reverse turns. 310-helices constitute nearly 10–15% of all helices in protein secondary structures, and are typically observed as extensions of α-helices found at either their N- or C- termini. Because of the α-helices tendency to consistently fold and unfold, it has been proposed that the 310-helix serves as an intermediary conformation of sorts, and provides insight into the initiation of α-helix folding.
A helix bundle is a small protein fold composed of several alpha helices that are usually nearly parallel or antiparallel to each other.

An alpha solenoid is a protein fold composed of repeating alpha helix subunits, commonly helix-turn-helix motifs, arranged in antiparallel fashion to form a superhelix. Alpha solenoids are known for their flexibility and plasticity. Like beta propellers, alpha solenoids are a form of solenoid protein domain commonly found in the proteins comprising the nuclear pore complex. They are also common in membrane coat proteins known as coatomers, such as clathrin, and in regulatory proteins that form extensive protein-protein interactions with their binding partners. Examples of alpha solenoid structures binding RNA and lipids have also been described.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.
Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. Pectate lyase is responsible for the eliminative cleavage of pectate, yielding oligosaccharides with 4-deoxy-α-D-mann-4-enuronosyl groups at their non-reducing ends. The protein is maximally expressed late in pollen development. It has been suggested that the pollen expression of pectate lyase genes might relate to a requirement for pectin degradation during pollen tube growth.

Pentapeptide repeats are a family of sequence motifs found in multiple tandem copies in protein molecules. Pentapeptide repeat proteins are found in all species, but they are found in many copies in cyanobacterial genomes. The repeats were first identified by Black and colleagues in the hglK protein. The later Bateman et al. showed that a large family of related pentapeptide repeat proteins existed. The function of these repeats is uncertain in most proteins. However, in the MfpA protein a DNA gyrase inhibitor it has been suggested that the pentapeptide repeat structure mimics the structure of DNA. The repeats form a regular right handed four sided beta helix structure known as the Rfr-fold.

In molecular biology, protein fold classes are broad categories of protein tertiary structure topology. They describe groups of proteins that share similar amino acid and secondary structure proportions. Each class contains multiple, independent protein superfamilies.
RiAFP refers to an antifreeze protein (AFP) produced by the Rhagium inquisitor longhorned beetle. It is a type V antifreeze protein with a molecular weight of 12.8 kDa; this type of AFP is noted for its hyperactivity. R. inquisitor is a freeze-avoidant species, meaning that, due to its AFP, R. inquisitor prevents its body fluids from freezing altogether. This contrasts with freeze-tolerant species, whose AFPs simply depress levels of ice crystal formation in low temperatures. Whereas most insect antifreeze proteins contain cysteines at least every sixth residue, as well as varying numbers of 12- or 13-mer repeats of 8.3-12.5kDa, RiAFP is notable for containing only one disulfide bridge. This property of RiAFP makes it particularly attractive for recombinant expression and biotechnological applications.

Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown.
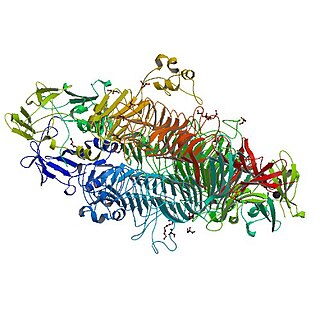
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.

The peridinin-chlorophyll-protein complex is a soluble molecular complex consisting of the peridinin-chlorophyll a-protein bound to peridinin, chlorophyll, and lipids. The peridinin molecules absorb light in the blue-green wavelengths and transfer energy to the chlorophyll molecules with extremely high efficiency. PCP complexes are found in many photosynthetic dinoflagellates, in which they may be the primary light-harvesting complexes.