
Analytical chemistry studies and uses instruments and methods to separate, identify, and quantify matter. In practice, separation, identification or quantification may constitute the entire analysis or be combined with another method. Separation isolates analytes. Qualitative analysis identifies analytes, while quantitative analysis determines the numerical amount or concentration.

Schellinger, Adam P.; Carr, Peter W. (2006). "Isocratic and gradient elution chromatography: A comparison in terms of speed, retention reproducibility and quantitation". Journal of Chromatography A. 19th International Symposium on MicroScale Bioseparations. 1109 (2): 253–266. doi:10.1016/j.chroma.2006.01.047. ISSN 0021-9673. PMID 16460742. S2CID 26072994.

Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a mass spectrum, a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures.

Electron ionization is an ionization method in which energetic electrons interact with solid or gas phase atoms or molecules to produce ions. EI was one of the first ionization techniques developed for mass spectrometry. However, this method is still a popular ionization technique. This technique is considered a hard ionization method, since it uses highly energetic electrons to produce ions. This leads to extensive fragmentation, which can be helpful for structure determination of unknown compounds. EI is the most useful for organic compounds which have a molecular weight below 600. Also, several other thermally stable and volatile compounds in solid, liquid and gas states can be detected with the use of this technique when coupled with various separation methods.

Gas chromatography–mass spectrometry (GC–MS) is an analytical method that combines the features of gas-chromatography and mass spectrometry to identify different substances within a test sample. Applications of GC–MS include drug detection, fire investigation, environmental analysis, explosives investigation, food and flavor analysis, and identification of unknown samples, including that of material samples obtained from planet Mars during probe missions as early as the 1970s. GC–MS can also be used in airport security to detect substances in luggage or on human beings. Additionally, it can identify trace elements in materials that were previously thought to have disintegrated beyond identification. Like liquid chromatography–mass spectrometry, it allows analysis and detection even of tiny amounts of a substance.

Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates, and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles. The metabolome represents the complete set of metabolites in a biological cell, tissue, organ, or organism, which are the end products of cellular processes. Messenger RNA (mRNA), gene expression data, and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism. There are indeed quantifiable correlations between the metabolome and the other cellular ensembles, which can be used to predict metabolite abundances in biological samples from, for example mRNA abundances. One of the ultimate challenges of systems biology is to integrate metabolomics with all other -omics information to provide a better understanding of cellular biology.

The metabolome refers to the complete set of small-molecule chemicals found within a biological sample. The biological sample can be a cell, a cellular organelle, an organ, a tissue, a tissue extract, a biofluid or an entire organism. The small molecule chemicals found in a given metabolome may include both endogenous metabolites that are naturally produced by an organism as well as exogenous chemicals that are not naturally produced by an organism.

Liquid chromatography–mass spectrometry (LC–MS) is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography with the mass analysis capabilities of mass spectrometry (MS). Coupled chromatography - MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides spectral information that may help to identify each separated component. MS is not only sensitive, but provides selective detection, relieving the need for complete chromatographic separation. LC–MS is also appropriate for metabolomics because of its good coverage of a wide range of chemicals. This tandem technique can be used to analyze biochemical, organic, and inorganic compounds commonly found in complex samples of environmental and biological origin. Therefore, LC–MS may be applied in a wide range of sectors including biotechnology, environment monitoring, food processing, and pharmaceutical, agrochemical, and cosmetic industries. Since the early 2000s, LC–MS has also begun to be used in clinical applications.
Protein methods are the techniques used to study proteins. There are experimental methods for studying proteins. Computational methods typically use computer programs to analyze proteins. However, many experimental methods require computational analysis of the raw data.

Atmospheric pressure chemical ionization (APCI) is an ionization method used in mass spectrometry which utilizes gas-phase ion-molecule reactions at atmospheric pressure (105 Pa), commonly coupled with high-performance liquid chromatography (HPLC). APCI is a soft ionization method similar to chemical ionization where primary ions are produced on a solvent spray. The main usage of APCI is for polar and relatively less polar thermally stable compounds with molecular weight less than 1500 Da. The application of APCI with HPLC has gained a large popularity in trace analysis detection such as steroids, pesticides and also in pharmacology for drug metabolites.

Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE), preparative one-dimensional gel electrophoresis, or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein identification, MS technology can identify and quantify the changes.

Two-dimensional chromatography is a type of chromatographic technique in which the injected sample is separated by passing through two different separation stages. Two different chromatographic columns are connected in sequence, and the effluent from the first system is transferred onto the second column. Typically the second column has a different separation mechanism, so that bands that are poorly resolved from the first column may be completely separated in the second column. Alternately, the two columns might run at different temperatures. During the second stage of separation the rate at which the separation occurs must be faster than the first stage, since there is still only a single detector. The plane surface is amenable to sequential development in two directions using two different solvents.

Capillary electrophoresis–mass spectrometry (CE–MS) is an analytical chemistry technique formed by the combination of the liquid separation process of capillary electrophoresis with mass spectrometry. CE–MS combines advantages of both CE and MS to provide high separation efficiency and molecular mass information in a single analysis. It has high resolving power and sensitivity, requires minimal volume and can analyze at high speed. Ions are typically formed by electrospray ionization, but they can also be formed by matrix-assisted laser desorption/ionization or other ionization techniques. It has applications in basic research in proteomics and quantitative analysis of biomolecules as well as in clinical medicine. Since its introduction in 1987, new developments and applications have made CE-MS a powerful separation and identification technique. Use of CE–MS has increased for protein and peptides analysis and other biomolecules. However, the development of online CE–MS is not without challenges. Understanding of CE, the interface setup, ionization technique and mass detection system is important to tackle problems while coupling capillary electrophoresis to mass spectrometry.

A triple quadrupole mass spectrometer (TQMS), is a tandem mass spectrometer consisting of two quadrupole mass analyzers in series, with a (non-mass-resolving) radio frequency (RF)–only quadrupole between them to act as a cell for collision-induced dissociation. This configuration is often abbreviated QqQ, here Q1q2Q3.
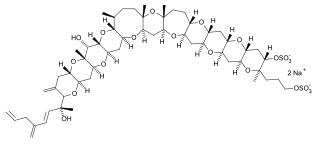
Yessotoxins are a group of lipophilic, sulfur bearing polyether toxins that are related to ciguatoxins. They are produced by a variety of dinoflagellates, most notably Lingulodinium polyedrum and Gonyaulax spinifera.

Instrumental analysis is a field of analytical chemistry that investigates analytes using scientific instruments.
Atmospheric pressure laser ionization is an atmospheric pressure ionization method for mass spectrometry (MS). Laser light in the UV range is used to ionize molecules in a resonance-enhanced multiphoton ionization (REMPI) process. It is a selective and sensitive ionization method for aromatic and polyaromatic compounds. Atmospheric photoionization is the latest in development of atmospheric ionization methods.
Mass spectrometric immunoassay (MSIA) is a rapid method is used to detect and/ or quantify antigens and or antibody analytes. This method uses an analyte affinity isolation to extract targeted molecules and internal standards from biological fluid in preparation for matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). This method allows for "top down" and "bottom up" analysis. This sensitive method allows for a new and improved process for detecting multiple antigens and antibodies in a single assay. This assay is also capable of distinguishing mass shifted forms of the same molecule via a panantibody, as well as distinguish point mutations in proteins. Each specific form is detected uniquely based on their characteristic molecular mass. MSIA has dual specificity because of the antibody-antigen reaction coupled with the power of a mass spectrometer.
Ion suppression in LC-MS and LC-MS/MS refers to reduced detector response, or signal:noise as a manifested effect of competition for ionisation efficiency in the ionisation source, between the analyte(s) of interest and other endogenous or exogenous species which have not been removed from the sample matrix during sample preparation. Ion suppression is not strictly a problem unless interfering compounds elute at the same time as the analyte of interest. In cases where ion suppressing species do co-elute with an analyte, the effects on the important analytical parameters including precision, accuracy and limit of detection can be extensive, severely limiting the validity of an assay's results.
Electro membrane extraction, or EME, is a miniaturized liquid-liquid extraction technique developed for sample preparation of aqueous samples prior to analysis by chromatography, electrophoresis, mass spectrometry, and related techniques in analytical chemistry. EME involves the use of a small supported liquid membrane (SLM) sustained in the wall of a porous hollow fiber, and application of an electrical field across the SLM.














