Related Research Articles

The human genome is a complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in a small DNA molecule found within individual mitochondria. These are usually treated separately as the nuclear genome and the mitochondrial genome. Human genomes include both protein-coding DNA sequences and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly repetitive sequences. Introns make up a large percentage of non-coding DNA. Some of this non-coding DNA is non-functional junk DNA, such as pseudogenes, but there is no firm consensus on the total amount of junk DNA.
In human genetics, the Mitochondrial Eve is the matrilineal most recent common ancestor (MRCA) of all living humans. In other words, she is defined as the most recent woman from whom all living humans descend in an unbroken line purely through their mothers and through the mothers of those mothers, back until all lines converge on one woman.
In human genetics, the Y-chromosomal most recent common ancestor is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent male from whom all living humans are descended through an unbroken line of their male ancestors. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve", the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct.
Archaeogenetics is the study of ancient DNA using various molecular genetic methods and DNA resources. This form of genetic analysis can be applied to human, animal, and plant specimens. Ancient DNA can be extracted from various fossilized specimens including bones, eggshells, and artificially preserved tissues in human and animal specimens. In plants, ancient DNA can be extracted from seeds and tissue. Archaeogenetics provides us with genetic evidence of ancient population group migrations, domestication events, and plant and animal evolution. The ancient DNA cross referenced with the DNA of relative modern genetic populations allows researchers to run comparison studies that provide a more complete analysis when ancient DNA is compromised.

In genetics and bioinformatics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome that is present in a sufficiently large fraction of considered population.
In biology and genetic genealogy, the most recent common ancestor (MRCA), also known as the last common ancestor (LCA), of a set of organisms is the most recent individual from which all the organisms of the set are descended. The term is also used in reference to the ancestry of groups of genes (haplotypes) rather than organisms.
Genetic genealogy is the use of genealogical DNA tests, i.e., DNA profiling and DNA testing, in combination with traditional genealogical methods, to infer genetic relationships between individuals. This application of genetics came to be used by family historians in the 21st century, as DNA tests became affordable. The tests have been promoted by amateur groups, such as surname study groups or regional genealogical groups, as well as research projects such as the Genographic Project.
Coalescent theory is a model of how alleles sampled from a population may have originated from a common ancestor. In the simplest case, coalescent theory assumes no recombination, no natural selection, and no gene flow or population structure, meaning that each variant is equally likely to have been passed from one generation to the next. The model looks backward in time, merging alleles into a single ancestral copy according to a random process in coalescence events. Under this model, the expected time between successive coalescence events increases almost exponentially back in time. Variance in the model comes from both the random passing of alleles from one generation to the next, and the random occurrence of mutations in these alleles.

Noah Aubrey Rosenberg is a geneticist working in evolutionary biology, mathematical phylogenetics, and population genetics, and is the Stanford Professor of Population Genetics and Society. His research focuses on mathematical modeling and statistical methods for genetics and evolution and he is the editor-in-chief of Theoretical Population Biology.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
David Emil Reich is an American geneticist known for his research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize.
Human evolutionary genetics studies how one human genome differs from another human genome, the evolutionary past that gave rise to the human genome, and its current effects. Differences between genomes have anthropological, medical, historical and forensic implications and applications. Genetic data can provide important insights into human evolution.

Guido Barbujani is an Italian population geneticist, evolutionary biologist and literary author born in Adria, who has worked with the State University of New York at Stony Brook (NY), University of Padua, and University of Bologna. He has taught at the University of Ferrara since 1996.

The 1000 Genomes Project, launched in January 2008, was an international research effort to establish by far the most detailed catalogue of human genetic variation. Scientists planned to sequence the genomes of at least one thousand anonymous participants from a number of different ethnic groups within the following three years, using newly developed technologies which were faster and less expensive. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal Nature. In 2012, the sequencing of 1092 genomes was announced in a Nature publication. In 2015, two papers in Nature reported results and the completion of the project and opportunities for future research.
In the context of the recent African origin of modern humans, the Southern Dispersal scenario refers to the early migration along the southern coast of Asia, from the Arabian Peninsula via Persia and India to Southeast Asia and Oceania. Alternative names include the "southern coastal route" or "rapid coastal settlement", with later descendants of those migrations eventually colonizing the rest of Eastern Eurasia, the remainder of Oceania, and the Americas.
The human mitochondrial molecular clock is the rate at which mutations have been accumulating in the mitochondrial genome of hominids during the course of human evolution. The archeological record of human activity from early periods in human prehistory is relatively limited and its interpretation has been controversial. Because of the uncertainties from the archeological record, scientists have turned to molecular dating techniques in order to refine the timeline of human evolution. A major goal of scientists in the field is to develop an accurate hominid mitochondrial molecular clock which could then be used to confidently date events that occurred during the course of human evolution.
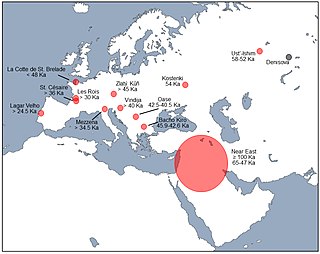
Interbreeding between archaic and modern humans occurred during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.
A ghost population is a population that has been inferred through using statistical techniques.
Sarah Anne Tishkoff is an American geneticist and the David and Lyn Silfen Professor in the Department of Genetics and Biology at the University of Pennsylvania. She also serves as a director for the American Society of Human Genetics and is an associate editor at PLOS Genetics, G3, and Genome Research. She is also a member of the scientific advisory board at the David and Lucile Packard Foundation.

The domestic cat originated from Near-Eastern and Egyptian populations of the African wildcat, Felis sylvestris lybica. The family Felidae, to which all living feline species belong, arose about ten to eleven million years ago and is divided into eight major phylogenetic lineages. The Felis lineage in particular is the lineage that the domestic cat is a member of. A number of investigations have shown that all domestic varieties of cats come from a single species of the Felis lineage, Felis catus. Variations of this lineage are found all over the world, and until recently scientists have had a hard time pinning down exactly which region gave rise to modern domestic cat breeds. Scientists believed that it was not just one incident that led to the domesticated cat but multiple, independent incidents at different places that led to these breeds. More complications arose from the fact that the wildcat population as a whole is very widespread and very similar to one another. These variations of wildcat can and will interbreed freely with one another when in close contact, further blurring the lines between taxa. Recent DNA studies, advancement in genetic technologies, and a better understanding of DNA and genetics as a whole has helped make discoveries in the evolutionary history of the domestic cat. Archaeological evidence has documented earlier dates of domestication than formerly believed.
References
- 1 2 3 Rotman, David (October 15, 2018). "DNA databases are too white. This man aims to fix that.: Carlos D. Bustamante's hunt for genetic variations between populations should help us better understand and treat disease". MIT Technology Review . Retrieved October 15, 2018.
- ↑ "Carlos Bustamante: Professor of Biomedical Data Science, of Genetics and, by courtesy, of Biology". Stanford University . Retrieved October 15, 2018.
Dr. Carlos D. Bustamante is an internationally recognized leader in the application of data science and genomics technology to problems in medicine, agriculture, and biology.
- 1 2 "Carlos D. Bustamante: Population Geneticist, Class of 2010". MacArthur Foundation . 2010. Retrieved October 15, 2018.
Carlos D. Bustamante is a population biologist who mines DNA sequence data for insights into the dynamics and migration of populations and the mechanisms of evolution and natural selection. In studies of humans, Bustamante analyzes SNPs (sites of common variation in a DNA sequence) from many individuals to infer changes in human populations and their relationship to specific gene mutations. He compared SNPs in regions of DNA that are translated into proteins with those in non-coding regions of the genome; from this analysis, he inferred that between a third and a half of mutations that change protein composition are lethal or produce weak negative selection, generating further understanding of a long-standing question of population genetics. He has applied SNP-based methods to retrace the history of species' domestication, both plants and animals; collaborative investigations of Asian rice and dogs, for example, have provided clues about where and how long ago humans domesticated these species. Bustamante has also teased out higher-resolution reconstructions of human demographic and migration patterns using new data sets from ethnically and geographically diverse samples. He and his colleagues have used DNA markers to assess the impact of shared language and geographic obstacles on migration patterns and genetic composition of human subpopulations in Europe, Africa, and Latin America. Through his multifaceted research, Bustamante is developing a rigorous, quantitative foundation for addressing fundamental questions about genetics and evolution across species, about patterns of population migration, and about the complex origins of human genetic diversity, before recorded history and since.
- ↑ Quenqua, Douglas (2013-08-12). "New Studies Suggest an 'Adam' and 'Eve' Link". The New York Times. ISSN 0362-4331 . Retrieved 2020-03-31.
- ↑ Linskey, Annie (October 15, 2018). "Warren releases results of DNA test". Boston Globe . Retrieved October 15, 2018.