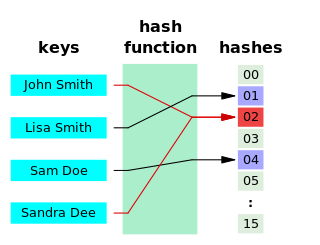
A hash function is any function that can be used to map data of arbitrary size to fixed-size values, though there are some hash functions that support variable length output. The values returned by a hash function are called hash values, hash codes, digests, or simply hashes. The values are usually used to index a fixed-size table called a hash table. Use of a hash function to index a hash table is called hashing or scatter storage addressing.

In computational complexity theory, NP is a complexity class used to classify decision problems. NP is the set of decision problems for which the problem instances, where the answer is "yes", have proofs verifiable in polynomial time by a deterministic Turing machine, or alternatively the set of problems that can be solved in polynomial time by a nondeterministic Turing machine.
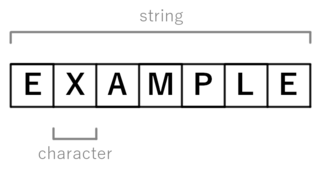
In computer programming, a string is traditionally a sequence of characters, either as a literal constant or as some kind of variable. The latter may allow its elements to be mutated and the length changed, or it may be fixed. A string is generally considered as a data type and is often implemented as an array data structure of bytes that stores a sequence of elements, typically characters, using some character encoding. String may also denote more general arrays or other sequence data types and structures.

In information theory, the Hamming distance between two strings or vectors of equal length is the number of positions at which the corresponding symbols are different. In other words, it measures the minimum number of substitutions required to change one string into the other, or equivalently, the minimum number of errors that could have transformed one string into the other. In a more general context, the Hamming distance is one of several string metrics for measuring the edit distance between two sequences. It is named after the American mathematician Richard Hamming.
In information theory, linguistics, and computer science, the Levenshtein distance is a string metric for measuring the difference between two sequences. Informally, the Levenshtein distance between two words is the minimum number of single-character edits required to change one word into the other. It is named after the Soviet mathematician Vladimir Levenshtein, who considered this distance in 1965.
In computational linguistics and computer science, edit distance is a string metric, i.e. a way of quantifying how dissimilar two strings are to one another, that is measured by counting the minimum number of operations required to transform one string into the other. Edit distances find applications in natural language processing, where automatic spelling correction can determine candidate corrections for a misspelled word by selecting words from a dictionary that have a low distance to the word in question. In bioinformatics, it can be used to quantify the similarity of DNA sequences, which can be viewed as strings of the letters A, C, G and T.
In computer science, parameterized complexity is a branch of computational complexity theory that focuses on classifying computational problems according to their inherent difficulty with respect to multiple parameters of the input or output. The complexity of a problem is then measured as a function of those parameters. This allows the classification of NP-hard problems on a finer scale than in the classical setting, where the complexity of a problem is only measured as a function of the number of bits in the input. This appears to have been first demonstrated in Gurevich, Stockmeyer & Vishkin (1984). The first systematic work on parameterized complexity was done by Downey & Fellows (1999).

In computer science, a suffix tree is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
In computer science, a longest common substring of two or more strings is a longest string that is a substring of all of them. There may be more than one longest common substring. Applications include data deduplication and plagiarism detection.
In computer science, the shortest common supersequence of two sequences X and Y is the shortest sequence which has X and Y as subsequences. This is a problem closely related to the longest common subsequence problem. Given two sequences X = < x1,...,xm > and Y = < y1,...,yn >, a sequence U = < u1,...,uk > is a common supersequence of X and Y if items can be removed from U to produce X and Y.
MAX-3SAT is a problem in the computational complexity subfield of computer science. It generalises the Boolean satisfiability problem (SAT) which is a decision problem considered in complexity theory. It is defined as:
In information theory and computer science, the Damerau–Levenshtein distance is a string metric for measuring the edit distance between two sequences. Informally, the Damerau–Levenshtein distance between two words is the minimum number of operations required to change one word into the other.

In computer science, approximate string matching is the technique of finding strings that match a pattern approximately. The problem of approximate string matching is typically divided into two sub-problems: finding approximate substring matches inside a given string and finding dictionary strings that match the pattern approximately.
String functions are used in computer programming languages to manipulate a string or query information about a string.
In theoretical computer science, a computational problem is a problem that may be solved by an algorithm. For example, the problem of factoring
In machine learning and data mining, a string kernel is a kernel function that operates on strings, i.e. finite sequences of symbols that need not be of the same length. String kernels can be intuitively understood as functions measuring the similarity of pairs of strings: the more similar two strings a and b are, the higher the value of a string kernel K(a, b) will be.
In computer science, the Wagner–Fischer algorithm is a dynamic programming algorithm that computes the edit distance between two strings of characters.
In computer science, the longest palindromic substring or longest symmetric factor problem is the problem of finding a maximum-length contiguous substring of a given string that is also a palindrome. For example, the longest palindromic substring of "bananas" is "anana". The longest palindromic substring is not guaranteed to be unique; for example, in the string "abracadabra", there is no palindromic substring with length greater than three, but there are two palindromic substrings with length three, namely, "aca" and "ada". In some applications it may be necessary to return all maximal palindromic substrings rather than returning only one substring or returning the maximum length of a palindromic substring.
In the field of computational biology, a planted motif search (PMS) also known as a (l, d)-motif search (LDMS) is a method for identifying conserved motifs within a set of nucleic acid or peptide sequences.
In mathematics, the Chvátal–Sankoff constants are mathematical constants that describe the lengths of longest common subsequences of random strings. Although the existence of these constants has been proven, their exact values are unknown. They are named after Václav Chvátal and David Sankoff, who began investigating them in the mid-1970s.







