Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and can occur in the digestive tracts of animals including ruminants and humans, where they are responsible for the methane content of belching and flatulence. In marine sediments, the biological production of methane, termed methanogenesis, is generally confined to where sulfates are depleted below the top layers. Methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Other methanogens are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface.
Methanogenesis or biomethanation is the formation of methane coupled to energy conservation by microbes known as methanogens. Organisms capable of producing methane for energy conservation have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic.

Methanosarcina is a genus of euryarchaeote archaea that produce methane. These single-celled organisms are known as anaerobic methanogens that produce methane using all three metabolic pathways for methanogenesis. They live in diverse environments where they can remain safe from the effects of oxygen, whether on the earth's surface, in groundwater, in deep sea vents, and in animal digestive tracts. Methanosarcina grow in colonies.
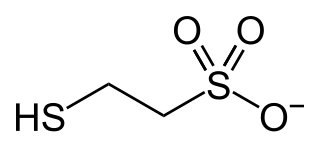
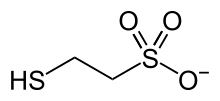
Coenzyme M is a coenzyme required for methyl-transfer reactions in the metabolism of archaeal methanogens, and in the metabolism of other substrates in bacteria. It is also a necessary cofactor in the metabolic pathway of alkene-oxidizing bacteria. CoM helps eliminate the toxic epoxides formed from the oxidation of alkenes such as propylene. The structure of this coenzyme was discovered by CD Taylor and RS Wolfe in 1974 while they were studying methanogenesis, the process by which carbon dioxide is transformed into methane in some anaerobic bacteria. The coenzyme is an anion with the formula HSCH
2CH
2SO−
3. It is named 2-mercaptoethanesulfonate and abbreviated HS–CoM. The cation is unimportant, but the sodium salt is most available. Mercaptoethanesulfonate contains both a thiol, which is the main site of reactivity, and a sulfonate group, which confers solubility in aqueous media.
Coenzyme B is a coenzyme required for redox reactions in methanogens. The full chemical name of coenzyme B is 7-mercaptoheptanoylthreoninephosphate. The molecule contains a thiol, which is its principal site of reaction.

The Wood–Ljungdahl pathway is a set of biochemical reactions used by some bacteria. It is also known as the reductive acetyl-coenzyme A (Acetyl-CoA) pathway. This pathway enables these organisms to use hydrogen as an electron donor, and carbon dioxide as an electron acceptor and as a building block for biosynthesis.
Anaerobic oxidation of methane (AOM) is a methane-consuming microbial process occurring in anoxic marine and freshwater sediments. AOM is known to occur among mesophiles, but also in psychrophiles, thermophiles, halophiles, acidophiles, and alkophiles. During AOM, methane is oxidized with different terminal electron acceptors such as sulfate, nitrate, nitrite and metals, either alone or in syntrophy with a partner organism.
In enzymology, a coenzyme F420 hydrogenase (EC 1.12.98.1) is an enzyme that catalyzes the chemical reaction
The enzyme sirohydrochlorin cobaltochelatase (EC 4.99.1.3) catalyzes the reaction

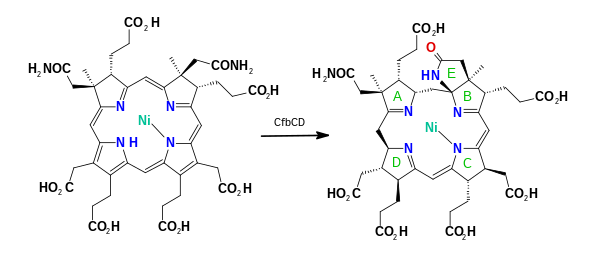
In enzymology, coenzyme-B sulfoethylthiotransferase, also known as methyl-coenzyme M reductase (MCR) or most systematically as 2-(methylthio)ethanesulfonate:N-(7-thioheptanoyl)-3-O-phosphothreonine S-(2-sulfoethyl)thiotransferase is an enzyme that catalyzes the final step in the formation of methane. It does so by combining the hydrogen donor coenzyme B and the methyl donor coenzyme M. Via this enzyme, most of the natural gas on earth was produced. Ruminants produce methane because their rumens contain methanogenic prokaryotes (Archaea) that encode and express the set of genes of this enzymatic complex.
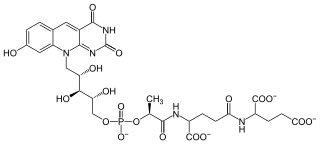
Coenzyme F420 or 8-hydroxy-5-deazaflavin is a coenzyme (sometimes called a cofactor) involved in redox reactions in methanogens, in many Actinomycetota, and sporadically in other bacterial lineages. It is a flavin derivative with an absorption maximum at 420 nm—hence its name. The coenzyme is a substrate for coenzyme F420 hydrogenase, 5,10-methylenetetrahydromethanopterin reductase and methylenetetrahydromethanopterin dehydrogenase.

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.
Methanocaldococcus jannaschii is a thermophilic methanogenic archaean in the class Methanococci. It was the first archaeon, and third organism, to have its complete genome sequenced. The sequencing identified many genes unique to the archaea. Many of the synthesis pathways for methanogenic cofactors were worked out biochemically in this organism, as were several other archaeal-specific metabolic pathways.

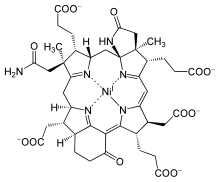
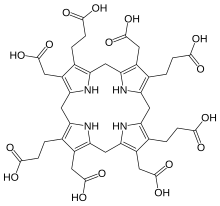
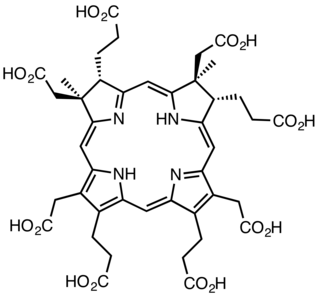
Sirohydrochlorin is a tetrapyrrole macrocyclic metabolic intermediate in the biosynthesis of sirohaem, the iron-containing prosthetic group in sulfite reductase enzymes. It is also the biosynthetic precursor to cofactor F430, an enzyme which catalyzes the release of methane in the final step of methanogenesis.

Dihydrosirohydrochlorin is one of several naturally occurring tetrapyrrole macrocyclic metabolic intermediates in the biosynthesis of vitamin B12 (cobalamin). Its oxidised form, sirohydrochlorin, is precursor to sirohaem, the iron-containing prosthetic group in sulfite reductase enzymes. Further biosynthetic transformations convert sirohydrochlorin to cofactor F430 for an enzyme which catalyzes the release of methane in the final step of methanogenesis.
In biochemistry, chelatases are enzymes that catalyze the insertion ("metalation") of naturally occurring tetrapyrroles. Many tetrapyrrole-based cofactors exist in nature including hemes, chlorophylls, and vitamin B12. These metallo cofactors are derived by the reaction of metal cations with tetrapyrroles, which are not ligands per se, but the conjugate acids thereof. In the case of ferrochelatases, the reaction that chelatases catalyze is:

C1 chemistry is the chemistry of one-carbon molecules. Although many compounds and ions contain only one carbon, stable and abundant C-1 feedstocks are the focus of research. Four compounds are of major industrial importance: methane, carbon monoxide, carbon dioxide, and methanol. Technologies that interconvert these species are often used massively to match supply to demand.
In enzymology, a formylmethanofuran dehydrogenase (EC 1.2.99.5) is an enzyme that catalyzes the chemical reaction:
Ralph Stoner Wolfe was an American microbiologist, who contributed to the discovery of the single-celled archaea as the third domain of life. He was a pioneer in the biochemistry of methanogenesis.

Hydroxyarchaeol is a core lipid unique to archaea, similar to archaeol, with a hydroxide functional group at the carbon-3 position of one of its ether side chains. It is found exclusively in certain taxa of methanogenic archaea, and is a common biomarker for methanogenesis and methane-oxidation. Isotopic analysis of hydroxyarchaeol can be informative about the environment and substrates for methanogenesis.