Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

Structural biology is a field that is many centuries old which, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization. Early structural biologists throughout the 19th and early 20th centuries were primarily only able to study structures to the limit of the naked eye's visual acuity and through magnifying glasses and light microscopes.

Protein folding is the physical process where a protein chain is translated into its native three-dimensional structure, typically a "folded" conformation, by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA into a linear chain of amino acids. At this stage, the polypeptide lacks any stable three-dimensional structure. As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure.

Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane. Peripheral membrane proteins are transiently associated with the cell membrane.
A kinemage is an interactive graphic scientific illustration. It often is used to visualize molecules, especially proteins although it can also represent other types of 3-dimensional data. The kinemage system is designed to optimize ease of use, interactive performance, and the perception and communication of detailed 3D information. The kinemage information is stored in a text file, human- and machine-readable, that describes the hierarchy of display objects and their properties, and includes optional explanatory text. The kinemage format is a defined chemical MIME type of 'chemical/x-kinemage' with the file extension '.kin'.

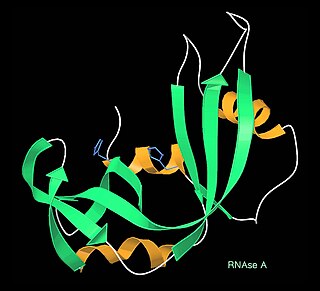
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a residue, which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo-electron microscopy (cryo-EM) and dual polarisation interferometry, to determine the structure of proteins.
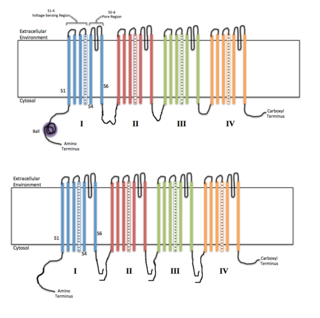
Voltage-gated ion channels are a class of transmembrane proteins that form ion channels that are activated by changes in the electrical membrane potential near the channel. The membrane potential alters the conformation of the channel proteins, regulating their opening and closing. Cell membranes are generally impermeable to ions, thus they must diffuse through the membrane through transmembrane protein channels. They have a crucial role in excitable cells such as neuronal and muscle tissues, allowing a rapid and co-ordinated depolarization in response to triggering voltage change. Found along the axon and at the synapse, voltage-gated ion channels directionally propagate electrical signals. Voltage-gated ion-channels are usually ion-specific, and channels specific to sodium (Na+), potassium (K+), calcium (Ca2+), and chloride (Cl−) ions have been identified. The opening and closing of the channels are triggered by changing ion concentration, and hence charge gradient, between the sides of the cell membrane.

Transmission electron cryomicroscopy (CryoTEM), commonly known as cryo-EM, is a form of cryogenic electron microscopy, more specifically a type of transmission electron microscopy (TEM) where the sample is studied at cryogenic temperatures. Cryo-EM, specifically 3-dimensional electron microscopy (3DEM), is gaining popularity in structural biology.
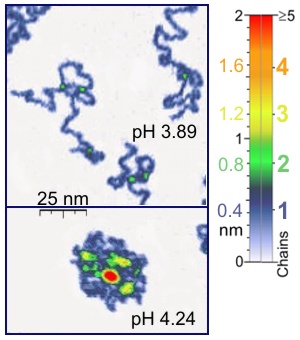
A single-molecule experiment is an experiment that investigates the properties of individual molecules. Single-molecule studies may be contrasted with measurements on an ensemble or bulk collection of molecules, where the individual behavior of molecules cannot be distinguished, and only average characteristics can be measured. Since many measurement techniques in biology, chemistry, and physics are not sensitive enough to observe single molecules, single-molecule fluorescence techniques caused a lot of excitement, since these supplied many new details on the measured processes that were not accessible in the past. Indeed, since the 1990s, many techniques for probing individual molecules have been developed.

In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins.
Nuclear magnetic resonance spectroscopy of proteins is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated.
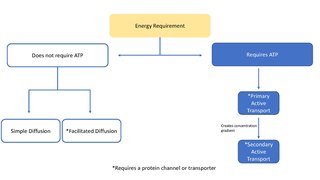
In biology, a transporter is a transmembrane protein that moves ions across a biological membrane to accomplish many different biological functions including, cellular communication, maintaining homeostasis, energy production, etc. There are different types of transporters including, pumps, uniporters, antiporters, and symporters. Active transporters or ion pumps are transporters that convert energy from various sources—including adenosine triphosphate (ATP), sunlight, and other redox reactions—to potential energy by pumping an ion up its concentration gradient. This potential energy could then be used by secondary transporters, including ion carriers and ion channels, to drive vital cellular processes, such as ATP synthesis.
Dual-polarization interferometry (DPI) is an analytical technique that probes molecular layers adsorbed to the surface of a waveguide using the evanescent wave of a laser beam. It is used to measure the conformational change in proteins, or other biomolecules, as they function.
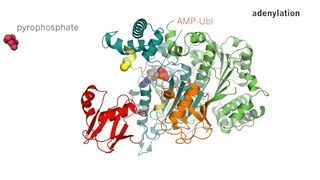
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.

Molecular biophysics is a rapidly evolving interdisciplinary area of research that combines concepts in physics, chemistry, engineering, mathematics and biology. It seeks to understand biomolecular systems and explain biological function in terms of molecular structure, structural organization, and dynamic behaviour at various levels of complexity. This discipline covers topics such as the measurement of molecular forces, molecular associations, allosteric interactions, Brownian motion, and cable theory. Additional areas of study can be found on Outline of Biophysics. The discipline has required development of specialized equipment and procedures capable of imaging and manipulating minute living structures, as well as novel experimental approaches.

Leadzyme is a small ribozyme (catalytic RNA), which catalyzes the cleavage of a specific phosphodiester bond. It was discovered using an in-vitro evolution study where the researchers were selecting for RNAs that specifically cleaved themselves in the presence of lead. However, since then, it has been discovered in several natural systems. Leadzyme was found to be efficient and dynamic in the presence of micromolar concentrations of lead ions. Unlike in other small self-cleaving ribozymes, other divalent metal ions cannot replace Pb2+ in the leadzyme. Due to obligatory requirement for a lead, the ribozyme is called a metalloribozyme.

Proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of conformations. Transitions between these states occur on a variety of length scales and time scales , and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis.
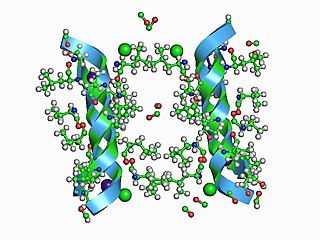
In electrophysiology, the term gating refers to the opening (activation) or closing of ion channels. This change in conformation is a response to changes in transmembrane voltage.

The term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular organelles and membranes and ribosomes, etc. that are complex mixtures of polypeptide, polynucleotide, polysaccharide or other polymeric macromolecules. They are generally of more than one of these types, and the mixtures are defined spatially, and with regard to their underlying chemical composition and structure. Macromolecules are found in living and nonliving things, and are composed of many hundreds or thousands of atoms held together by covalent bonds; they are often characterized by repeating units. Assemblies of these can likewise be biologic or non-biologic, though the MA term is more commonly applied in biology, and the term supramolecular assembly is more often applied in non-biologic contexts. MAs of macromolecules are held in their defined forms by non-covalent intermolecular interactions, and can be in either non-repeating structures, or in repeating linear, circular, spiral, or other patterns. The process by which MAs are formed has been termed molecular self-assembly, a term especially applied in non-biologic contexts. A wide variety of physical/biophysical, chemical/biochemical, and computational methods exist for the study of MA; given the scale of MAs, efforts to elaborate their composition and structure and discern mechanisms underlying their functions are at the forefront of modern structure science.
The Cluster of Excellence Frankfurt "Macromolecular Complexes" (CEF) was established in 2006 by Goethe University Frankfurt together with the Max Planck Institute of Biophysics and the Max Planck Institute for Brain Research in the context of the German Universities Excellence Initiative. Funding by the Deutsche Forschungsgemeinschaft (DFG) endet in October 2019. CEF grew out of the long-standing collaborative research on membrane proteins and RNA molecules and strengthened research efforts in these fields by recruiting further scientists to Frankfurt/Main. CEF brought together the research activities of up to 45 research groups, the majority of which were based on Riedberg Campus in Frankfurt/Main. CEF founded the Buchmann Institute for Molecular Life Sciences (BMLS).